BGQ
| Blue Gene/Q (BGQ) | |
|---|---|
| Installed | Aug 2012, Nov 2014 |
| Operating System | RH6.3, CNK (Linux) |
| Number of Nodes | 4096 nodes (65,536 cores) |
| Interconnect | 5D Torus (jobs), QDR Infiniband (I/O) |
| Ram/Node | 16 GB |
| Cores/Node | 16 (64 threads) |
| Login/Devel Node | bgqdev-fen1 |
| Vendor Compilers | bgxlc, bgxlf |
| Queue Submission | Loadleveler |
System Status
The current BGQ system status can be found on the wiki's Main Page.
SOSCIP & LKSAVI
The BGQ is a Southern Ontario Smart Computing Innovation Platform (SOSCIP) BlueGene/Q supercomputer located at the University of Toronto's SciNet HPC facility. The SOSCIP multi-university/industry consortium is funded by the Ontario Government and the Federal Economic Development Agency for Southern Ontario [1].
A half-rack of BlueGene/Q (8,192 cores) was purchased by the Li Ka Shing Institute of Virology at the University of Alberta in late fall 2014 and integrated into the existing BGQ system.
The combined 4 rack system is the fastest Canadian supercomputer on the top 500, currently at the 120th place (Nov 2015) -- 499th as of Aug 2018.
Support Email
Please use <bgq-support@scinet.utoronto.ca> for BGQ-specific inquiries.
Specifications
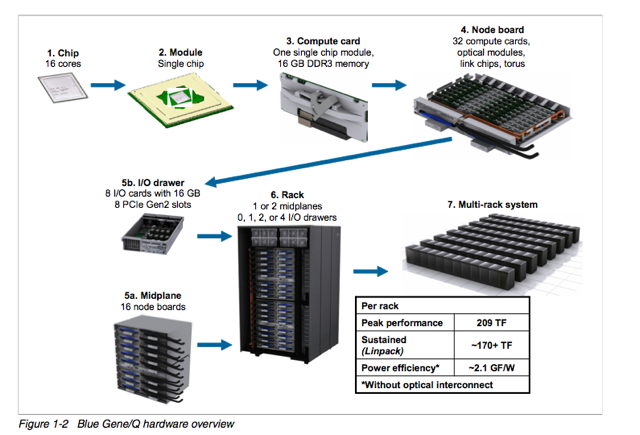
BGQ is an extremely dense and energy efficient 3rd generation Blue Gene IBM supercomputer built around a system-on-a-chip compute node that has a 16core 1.6GHz PowerPC based CPU (PowerPC A2) with 16GB of Ram. The nodes are bundled in groups of 32 into a node board (512 cores), and 16 boards make up a midplane (8192 cores) with 2 midplanes per rack, or 16,348 cores and 16 TB of RAM per rack. The compute nodes run a very lightweight Linux-based operating system called CNK (Compute Node Kernel). The compute nodes are all connected together using a custom 5D torus highspeed interconnect. Each rack has 16 I/O nodes that run a full Redhat Linux OS that manages the compute nodes and mounts the filesystem. SciNet's BGQ consists of 8 mdiplanes (four-racks) totalling 65,536 cores and 64TB of RAM.
5D Torus Network
The network topology of BlueGene/Q is a five-dimensional (5D) torus, with direct links between the nearest neighbours in the ±A, ±B, ±C, ±D, and ±E directions. As such there are only a few optimum block sizes that will use the network efficiently.
| Node Boards | Compute Nodes | Cores | Torus Dimensions |
| 1 | 32 | 512 | 2x2x2x2x2 |
| 2 (adjacent pairs) | 64 | 1024 | 2x2x4x2x2 |
| 4 (quadrants) | 128 | 2048 | 2x2x4x4x2 |
| 8 (halves) | 256 | 4096 | 4x2x4x4x2 |
| 16 (midplane) | 512 | 8192 | 4x4x4x4x2 |
| 32 (1 rack) | 1024 | 16384 | 4x4x4x8x2 |
| 64 (2 racks) | 2048 | 32768 | 4x4x8x8x2 |
| 96 (3 racks) | 3072 | 49152 | 4x4x12x8x2 |
| 128 (4 racks) | 4096 | 65536 | 8x4x8x8x2 |
On a 5D torus topology, each node has 10 point-to-point direct links. There is an additional 11th link to the I/O nodes.
Login/Devel Node
The development node is bgqdev-fen1 which one can login to directly from outside using bgqdev.scinet.utoronto.ca, e.g.
$ ssh -l USERNAME bgqdev.scinet.utoronto.ca -X
where USERNAME is your username on the BGQ and the -X flag is optional, needed only if you will use X graphics.
Note: To learn how to setup ssh keys for logging in please see SSH keys.
These development node is a Power7 machines running Linux which serve as the compilation and submission host for the BGQ. Programs are cross-compiled for the BGQ on this node and then submitted to the queue using loadleveler.
Modules and Environment Variables
To use most packages on the SciNet machines - including most of the compilers - , you will have to use the `modules' command. The command module load some-package will set your environment variables (PATH, LD_LIBRARY_PATH, etc) to include the default version of that package. module load some-package/specific-version will load a specific version of that package. This makes it very easy for different users to use different versions of compilers, MPI versions, libraries etc.
A list of the installed software can be seen on the system by typing
$ module avail
To load a module (for example, the default version of the intel compilers)
$ module load vacpp
To unload a module
$ module unload vacpp
To unload all modules
$ module purge
These commands can go in your .bashrc files to make sure you are using the correct packages.
Modules that load libraries, define environment variables pointing to the location of library files and include files for use Makefiles. These environment variables follow the naming convention
$SCINET_[short-module-name]_BASE $SCINET_[short-module-name]_LIB $SCINET_[short-module-name]_INC
for the base location of the module's files, the location of the libraries binaries and the header files, respectively.
So to compile and link the library, you will have to add -I${SCINET_[module-basename]_INC} and -L${SCINET_[module-basename]_LIB}, respectively, in addition to the usual -l[libname].
Note that a module load command only sets the environment variables in your current shell (and any subprocesses that the shell launches). It does not effect other shell environments.
If you always require the same modules, it is easiest to load those modules in your .bashrc and then they will always be present in your environment; if you routinely have to flip back and forth between modules, it is easiest to have almost no modules loaded in your .bashrc and simply load them as you need them (and have the required module load commands in your job submission scripts).
Compilers
The BGQ uses IBM XL compilers to cross-compile code for the BGQ. Compilers are available for FORTRAN, C, and C++. They are accessible by default, or by loading the xlf and vacpp modules. The compilers by default produce static binaries, however with BGQ it is possible to now use dynamic libraries as well. The compilers follow the XL conventions with the prefix bg, so bgxlc and bgxlf90 are the C and FORTRAN compilers respectively.
Most users however will use the MPI variants, i.e. mpixlf90 and mpixlc and which are available by loading the mpich2 module.
module load mpich2
It is recommended to use at least the following flags when compiling and linking
-O3 -qarch=qp -qtune=qp
If you want to build a package for which the configure script tries to run small test jobs, the cross-compiling nature of the bgq can get in the way. In that case, you should use the interactive debugjob environment as described below.
ION/Devel Nodes
There are also bgq native development nodes named bgqdev-ion[01-24] which one can login to directly, i.e. ssh, from bgqdev-fen1. These nodes are extra I/O nodes that are essentially the same as the BGQ compute nodes with the exception that they run a full RedHat Linux and have an infiniband interface providing direct network access. Unlike the regular development node, bgqdev-fen1, which is Power7, this node has the same BGQ A2 processor, and thus cross compilations are not required which can make building some software easier.
NOTE: BGQ MPI jobs can be compiled on these nodes, however can not be run locally as the mpich2 is setup for the BGQ network and thus will fail on these nodes.
Job Submission
As the BlueGene/Q architecture is different from the development nodes, you cannot run applications intended/compiled for the BGQ on the devel nodes. The only way to run (or even test) your program is to submit a job to the BGQ. Jobs are submitted as scripts through loadleveler. That script must then use runjob to start the job, which in many ways similar to mpirun or mpiexec. As shown above in the network topology overview, there are only a few optimum job size configurations which is also further constrained by each block requiring a minimum of one IO node. In SciNet's configuration (with 8 I/O nodes per midplane) this allows 64 nodes (1024 cores) to be the smallest block size. Normally a block size matches the job size to offer fully dedicated resources to the job. Smaller jobs can be run within the same block however this results in shared resources (network and IO) and are referred to as sub-block jobs and are described in more detail below.
runjob
All BGQ runs are launched using runjob which for those familiar with MPI is analogous to mpirun/mpiexec. Jobs run on a block, which is a predefined group of nodes that have already been configured and booted. There are two ways to get a block. One way is to use a 30-minute 'debugjob' session (more about that below). The other, more common case, is using a job script submitted and are running using loadleveler. Inside the job script, this block is set for you, and you do not have to specify the block name. For example, if your loadleveler job script requests 64 nodes, each with 16 cores (for a total of 1024 cores), from within that job script, you can run a job with 16 processes per node and 1024 total processes with
runjob --np 1024 --ranks-per-node=16 --cwd=$PWD : $PWD/code -f file.in
Here, --np 1024 sets the total number of mpi tasks, while --ranks-per-node=16 specifies that 16 processes should run on each node. For pure mpi jobs, it is advisable always to give the number of ranks per node, because the default value of 1 may leave 15 cores on the node idle. The argument to ranks-per-node may be 1, 2, 4, 8, 16, 32, or 64.
runjob flags are shown with
runjob -h
a particularly useful one is
--verbose #
where # is from 1-7 which can be helpful in debugging an application.
How to set ranks-per-node
There are 16 cores per node, but the argument to ranks-per-node may be 1, 2, 4, 8, 16, 32, or 64. While it may seem natural to set ranks-per-node to 16, this is not generally recommended. On the BGQ, one can efficiently run more than 1 process per core, because each core has four "hardware threads" (similar to HyperThreading on the GPC and Simultaneous Multi Threading on the TCS and P7), which can keep the different parts of each core busy at the same time. One would therefore ideally use 64 ranks per node. There are two main reason why one might not set ranks-per-node to 64:
- The memory requirements do not allow 64 ranks (each rank only has 256MB of memory)
- The application is more efficient in a hybrid MPI/OpenMP mode (or MPI/pthreads). Using less ranks-per-node, the hardware threads are used as OpenMP threads within each process.
Because threads can share memory, the memory requirements of the hybrid runs is typically smaller than that of pure MPI runs.
Note that the total number of mpi processes in a runjob (i.e., the --np argument) should be the ranks-per-node times the number of nodes (set by bg_size in the loadleveler script). So for the same number of nodes, if you change ranks-per-node by a factor of two, you should also multiply the total number of mpi processes by two.
Queue Limits
The maximum wall_clock_limit is 24 hours. Official SOSCIP project jobs are prioritized over all other jobs using a fairshare algorithm with a 14 day rolling window.
A 64 node block is reserved for development and interactive testing for 16 hours, from 8AM to midnight, everyday including weekends. While you can still reserve an interactive block from midnight to 8AM, the priority is given to batch jobs at that time interval in order to keep the machine usage as high as possible. This block is accessed by using the debugjob command which has a 30 minute maximum wall_clock_limit. The purpose of this reservation is to ensure short testing jobs are run quickly without being held up by longer production type jobs.
Batch Jobs
Job submission is done through loadleveler with a few blue gene specific commands. The command "bg_size" is in number of nodes, not cores, so a bg_size=64 would be 64x16=1024 cores.
The parameter bg_size can only be equal to 64, 128, 256, 512, 1024 and 2048.
np ≤ ranks-per-node * bg_size
ranks-per-node ≤ np
(ranks-per-node * OMP_NUM_THREADS ) ≤ 64
np : number of MPI processes
ranks-per-node : number of MPI processes per node = 1 , 2 , 4 , 8 , 16 , 32 , 64
OMP_NUM_THREADS : number of OpenMP thread per MPI process (for hybrid codes) = 1 , 2 , 4 , 8 , 16 , 32 , 64
#!/bin/sh # @ job_name = bgsample # @ job_type = bluegene # @ comment = "BGQ Job By Size" # @ error = $(job_name).$(Host).$(jobid).err # @ output = $(job_name).$(Host).$(jobid).out # @ bg_size = 64 # @ wall_clock_limit = 30:00 # @ bg_connectivity = Torus # @ queue # Launch all BGQ jobs using runjob runjob --np 1024 --ranks-per-node=16 --envs OMP_NUM_THREADS=1 --cwd=$SCRATCH/ : $HOME/mycode.exe myflags
To submit to the queue use
llsubmit myscript.sh
Steps ( Job dependency)
LoadLeveler has a lot of advanced features to control job submission and execution. One of these features is called steps. This feature allows a series of jobs to be submitted using one script with dependencies defined between the jobs. What this allows is for a series of jobs to be run sequentially, waiting for the previous job, called a step, to be finished before the next job is started. The following example uses the same LoadLeveler script as previously shown, however the #@ step_name and #@ dependency directives are used to rerun the same case three times in a row, waiting until each job is finished to start the next.
#!/bin/sh
# @ job_name = bgsample
# @ job_type = bluegene
# @ comment = "BGQ Job By Size"
# @ error = $(job_name).$(Host).$(jobid).err
# @ output = $(job_name).$(Host).$(jobid).out
# @ bg_size = 64
# @ wall_clock_limit = 30:00
# @ bg_connectivity = Torus
# @ step_name = step1
# @ queue
# Launch the first step :
if [ $LOADL_STEP_NAME = "step1" ]; then
runjob --np 1024 --ranks-per-node=16 --envs OMP_NUM_THREADS=1 --cwd=$SCRATCH/ : $HOME/mycode.exe myflags
fi
# @ job_name = bgsample
# @ job_type = bluegene
# @ comment = "BGQ Job By Size"
# @ error = $(job_name).$(Host).$(jobid).err
# @ output = $(job_name).$(Host).$(jobid).out
# @ bg_size = 64
# @ wall_clock_limit = 30:00
# @ bg_connectivity = Torus
# @ step_name = step2
# @ dependency = step1 == 0
# @ queue
# Launch the second step if the first one has returned 0 (done successfully) :
if [ $LOADL_STEP_NAME = "step2" ]; then
runjob --np 1024 --ranks-per-node=16 --envs OMP_NUM_THREADS=1 --cwd=$SCRATCH/ : $HOME/mycode.exe myflags
fi
# @ job_name = bgsample
# @ job_type = bluegene
# @ comment = "BGQ Job By Size"
# @ error = $(job_name).$(Host).$(jobid).err
# @ output = $(job_name).$(Host).$(jobid).out
# @ bg_size = 64
# @ wall_clock_limit = 30:00
# @ bg_connectivity = Torus
# @ step_name = step3
# @ dependency = step2 == 0
# @ queue
# Launch the third step if the second one has returned 0 (done successfully) :
if [ $LOADL_STEP_NAME = "step3" ]; then
runjob --np 1024 --ranks-per-node=16 --envs OMP_NUM_THREADS=1 --cwd=$SCRATCH/ : $HOME/mycode.exe myflags
fi
Monitoring Jobs
To see running jobs
llq2
or
llq -b
to cancel a job use
llcancel JOBID
and to look at details of the bluegene resources use
llbgstatus -M all
Note: the loadleveler script commands are not run on a bgq compute node but on the front-end node. Only programs started with runjob run on the bgq compute nodes. You should therefore keep scripting in the submission script to a bare minimum.
Monitoring Stats
Use llbgstats to monitor your own stats and/or your group stats. PIs can also print their (current) monthly report.
llbgstats -h
Interactive Use / Debugging
As BGQ codes are cross-compiled they cannot be run direclty on the front-nodes. Users however only have access to the BGQ through loadleveler which is appropriate for batch jobs, however an interactive session is typically beneficial when debugging and developing. As such a script has been written to allow a session in which runjob can be run interactively. The script uses loadleveler to setup a block and set all the correct environment variables and then launch a spawned shell on the front-end node. The debugjob session currently allows a 30 minute session on 64 nodes and when run on bgqdev runs in a dedicated reservation as described previously in the queue limits section.
[user@bgqdev-fen1]$ debugjob [user@bgqdev-fen1]$ runjob --np 64 --ranks-per-node=16 --cwd=$PWD : $PWD/my_code -f myflags [user@bgqdev-fen1]$ exit
For debugging, gdb and Allinea DDT are available. The latter is recommended as it automatically attaches to all the processes of a process (instead of attaching a gdbtool by hand (as explained in the BGQ Application Development guide, link below). Simply compile with -g, load the ddt/4.1 module, type ddt and follow the graphical user interface. The DDT user guide can be found below.
Note: when running a job under ddt, you'll need to add "--ranks-per-node=X" to the "runjob arguments" field.
Apart from debugging, this environment is also useful for building libraries and applications that need to run small tests as part of their 'configure' step. Within the debugjob session, applications compiled with the bgxl compilers or the mpcc/mpCC/mpfort wrappers, will automatically run on the BGQ, skipping the need for the runjob command, provided if you set the following environment variables
$ export BG_PGM_LAUNCHER=yes $ export RUNJOB_NP=1
The latter setting sets the number of mpi processes to run. Most configure scripts expect only one mpi process, thus, RUNJOB_NP=1 is appropriate.
debugjob session with an executable implicitly calls runjob with 1 mpi task :
debugjob -i
**********************************************************
Interactive BGQ runjob shell using bgq-fen1-ib0.10295.0 and
LL14040718574824 for 30 minutes with 64 NODES (1024 cores).
IMPLICIT MODE: running an executable implicitly calls runjob
with 1 mpi task
Exit shell when finished.
**********************************************************
Sub-block jobs
BGQ allows multiple applications to share the same block, which is referred to as sub-block jobs, however this needs to be done from within the same loadleveler submission script using multiple calls to runjob. To run a sub-block job, you need to specify a "--corner" within the block to start each job and a 5D Torus AxBxCxDxE "--shape". The starting corner will depend on the specific block details provided by loadleveler and the shape and size of job trying to be used.
Figuring out what the corners and shapes should be is very tricky (especially since it depends on the block you get allocated). For that reason, we've created a script called subblocks that determines the corners and shape of the sub-blocks. It only handles the (presumable common) case in which you want to subdivide the block into n equally sized sub-blocks, where n may be 1,2,4,8,16 and 32.
Here is an example script calling subblocks with a size of 4 that will return the appropriate $SHAPE argument and an array of 16 starting $CORNER.
#!/bin/bash
# @ job_name = bgsubblock
# @ job_type = bluegene
# @ comment = "BGQ Job SUBBLOCK "
# @ error = $(job_name).$(Host).$(jobid).err
# @ output = $(job_name).$(Host).$(jobid).out
# @ bg_size = 64
# @ wall_clock_limit = 30:00
# @ bg_connectivity = Torus
# @ queue
# Using subblocks script to set $SHAPE and array of ${CORNERS[n]}
# with size of subblocks in nodes (ie similiar to bg_size)
# In this case 16 sub-blocks of 4 cnodes each (64 total ie bg_size)
source subblocks 4
# 16 jobs of 4 each
for (( i=0; i < 16 ; i++)); do
runjob --corner ${CORNER[$i]} --shape $SHAPE --np 64 --ranks-per-node=16 : your_code_here > $i.out &
done
wait
Remember that subjobs are not the ideal way to run on the BlueGene/Qs. One needs to consider that these sub-blocks all have to share the same I/O nodes, so for I/O intensive jobs this will be an inefficient setup. Also consider that if you need to run such small jobs that you have to run in sub-blocks, it may be more efficient to use other clusters such as the GPC.
Let us know if you run into any issues with this technique, please contact bgq-support for help.
Filesystem
The BGQ has its own dedicated 500TB file system based on GPFS (General Parallel File System). There are two main systems for user data: /home, a small, backed-up space where user home directories are located, and /scratch, a large system for input or output data for jobs; data on /scratch is not backed up. The path to your home directory is in the environment variable $HOME, and will look like /home/G/GROUP/USER, . The path to your scratch directory is in the environment variable $SCRATCH, and will look like /scratch/G/GROUP/USER (following the conventions of the rest of the SciNet systems).
| file system | purpose | user quota | backed up | purged |
|---|---|---|---|---|
| /home | development | 50 GB | yes | never |
| /scratch | computation | first of (20 TB ; 1 million files) | no | not currently |
Transfering files
The BGQ GPFS file system, except for HPSS, is not shared with the other SciNet systems (gpc, tcs, p7, arc), nor is the other file system mounted on the BGQ. Use scp to copy files from one file system to the other, e.g., from bgqdev-fen1, you could do
$ scp -c arcfour login.scinet.utoronto.ca:code.tgz .
or from a login node you could do
$ scp -c arcfour code.tgz bgqdev.scinet.utoronto.ca:
The flag -c arcfour is optional. It tells scp (or really, ssh), to use a non-default encryption. The one chosen here, arcfour, has been found to speed up the transfer by a factor of two (you may expect around 85MB/s). This encryption method is only recommended for copying from the BGQ file system to the regular SciNet GPFS file system or back.
Note that although these transfers are witihin the same data center, you have to use the full names of the systems, login.scinet.utoronto.ca and bgq.scinet.utoronto.ca, respectively, and that you will be asked you for your password.
How much Disk Space Do I have left?
The diskUsage command, available on the bgqdev nodes, provides information in a number of ways on the home and scratch file systems. For instance, how much disk space is being used by yourself and your group (with the -a option), or how much your usage has changed over a certain period ("delta information") or you may generate plots of your usage over time. Please see the usage help below for more details.
Usage: diskUsage [-h|-?| [-a] [-u <user>] [-de|-plot]
-h|-?: help
-a: list usages of all members on the group
-u <user>: as another user on your group
-de: include delta information
-plot: create plots of disk usages
Note that the information on usage and quota is only updated hourly!
Bridge to HPSS
BGQ users may transfer material to/from HPSS via the GPC archive queue. On the HPSS gateway node (gpc-archive01), the BGQ GPFS file systems are mounted under a single mounting point /bgq (/bgq/scratch and /bgq/home). For detailed information on the use of HPSS please read the HPSS wiki section.
Software modules installed on the BGQ
| Software | Version | Comments | Command/Library | Module Name |
|---|---|---|---|---|
| Compilers & Development Tools | ||||
| IBM fortran compiler | 14.1 | These are cross compilers | bgxlf,bgxlf_r,bgxlf90,... | xlf |
| IBM c/c++ compilers | 12.1 | These are cross compilers | bgxlc,bgxlC,bgxlc_r,bgxlC_r,... | vacpp |
| MPICH2 MPI library | 1.4.1 | There are 4 versions (see BGQ Applications Development document). | mpicc,mpicxx,mpif77,mpif90 | mpich2 |
| GCC Compiler | 4.4.6, 4.8.1 | GNU Compiler Collection for BGQ (4.8.1 requires binutils/2.23 to be loaded) |
powerpc64-bgq-linux-gcc, powerpc64-bgq-linux-g++, powerpc64-bgq-linux-gfortran | bgqgcc |
| Clang Compiler | r217688-20140912, r263698-20160317 | Clang cross-compilers for bgq | powerpc64-bgq-linux-clang, powerpc64-bgq-linux-clang++ | bgclang |
| Binutils | 2.21.1, 2.23 | Cross-compilation utilities | addr2line, ar, ld, ... | binutils |
| CMake | 2.8.8, 2.8.12.1 | cross-platform, open-source build system | cmake | cmake |
| Git | 1.9.5 | Revision control system | git, gitk | git |
| Debug/performance tools | ||||
| gdb | 7.2 | GNU Debugger | gdb | gdb |
| ddd | 3.3.12 | GNO Data Display Debugger | ddd | ddd |
| DDT | 4.1, 4.2, 5.0.1 | Allinea's Distributed Debugging Tool | ddt | ddt |
| HPCTW | 1.0 | BGQ MPI and Hardware Counters | libmpihpm.a, libmpihpm_smp.a, libmpitrace.a | hptibm |
| MemP | 1.0.3 | BGQ Memory Stats | libmemP.a | memP |
| Storage tools/libraries | ||||
| HDF5 | 1.8.9-v18 | Scientific data storage and retrieval | h5ls, h5diff, ..., libhdf5 | hdf5/189-v18-serial-xlc* hdf5/189-v18-mpich2-xlc |
| HDF5 | 1.8.12-v18 | Scientific data storage and retrieval | h5ls, h5diff, ..., libhdf5 | hdf5/1812-v18-serial-gcc hdf5/1812-v18-mpich2-gcc |
| NetCDF | 4.2.1.1 | Scientific data storage and retrieval | ncdump,ncgen,libnetcdf | netcdf/4.2.1.1-serial-xlc* netcdf/4.2.1.1-mpich2-xlc |
| Parallel NetCDF | 1.3.1 | Parallel scientific data storage and retrieval using MPI-IO | libpnetcdf.a | parallel-netcdf |
| Libraries | ||||
| ESSL | 5.1 | IBM Engineering and Scientific Subroutine Library (manual below) | libesslbg,libesslsmpbg | essl |
| WSMP | 15.06.01 | Watson Sparse Matrix Package | libpwsmpBGQ.a | WSMP |
| FFTW | 2.1.5, 3.3.2, 3.1.2-esslwrapper | Fast fourier transform | libsfftw,libdfftw,libfftw3, libfftw3f | fftw/2.1.5, fftw/3.3.2, fftw/3.1.2-esslwrapper |
| LAPACK + ScaLAPACK | 3.4.2 + 2.0.2 | Linear algebra routines. A subset of Lapack may be found in ESSL as well. | liblapack, libscalpack | lapack |
| GSL | 1.15 | GNU Scientific Library | libgsl, libgslcblas | gsl |
| BOOST | 1.47.0, 1.54, 1.57 | C++ Boost libraries | libboost... | cxxlibraries/boost |
| bzip2 + szip + zlib | 1.0.6 + 2.1 + 1.2.7 | compression libraries | libbz2,libz,libsz | compression |
| METIS | 5.0.2 | Serial Graph Partitioning and Fill-reducing Matrix Ordering | libmetis | metis |
| ParMETIS | 4.0.2 | Parallel graph partitioning and fill-reducing matrix ordering | libparmetis | parmetis |
| OpenSSL | 1.0.2 | General-purpose cryptography library | libcrypto, libssl | openssl |
| FILTLAN | 1.0 | The Filtered Lanczos Package | libdfiltlan,libdmatkit,libsfiltlan,libsmatkit | FILTLAN |
| Scripting/interpreted languages | ||||
| Python | 2.6.6 | Python programming language | /bgsys/tools/Python-2.6/bin/python | python |
| Python | 2.7.3 | Python programming language. Modules included : numpy-1.8.0, pyFFTW-0.9.2, astropy-0.3, scipy-0.13.3, mpi4py-1.3.1, h5py-2.2.1 | /scinet/bgq/tools/Python/python2.7.3-20131205/bin/python | python |
| Python | 3.2.2 | Python programming language | /bgsys/tools/Python-3.2/bin/python3 | python |
| Applications | ||||
| ABINIT | 7.10.4 | An atomic-scale simulation software suite | abinit | abinit |
| BerkeleyGW library | 1.0.4-2.0.0436 | Computes quasiparticle properties and the optical responses of a large variety of materials | libBGW_wfn.a, wfn_rho_vxc_io_m.mod | BGW-paratec |
| CP2K | 2.3, 2.4, 2.5.1, 2.6.1 | DFT molecular dynamics, MPI | cp2k.psmp | cp2k |
| CPMD | 3.15.3, 3.17.1 | Carr-Parinello molecular dynamics, MPI | cpmd.x | cpmd |
| gnuplot | 4.6.1 | interactive plotting program to be run on front-end nodes | gnuplot | gnuplot |
| LAMMPS | Nov 2012/7Dec15/7Dec15-mpi | Molecular Dynamics | lmp_bgq | lammps |
| NAMD | 2.9 | Molecular Dynamics | namd2 | namd/2.9-smp |
| Quantum Espresso | 5.0.3/5.2.1 | Molecular Structure / Quantum Chemistry | qe_pw.x, etc | espresso |
| OpenFOAM | 2.2.0, 2.3.0, 2.4.0, 3.0.1, 5.0 | Computational Fluid Dynamics | icofoam,etc. | openfoam/2.2.0, openfoam/2.3.0, openfoam/2.4.0, openfoam/3.0.1, openfoam/5.0 |
| Beta Tests | ||||
| WATSON API | beta | Natural Language Processing | watson_beta | FEN/WATSON |
OpenFOAM on BGQ
A detailed explanation of OpenFOAM usage on the BG/Q cluster.
Python on BlueGene
Python 2.7.3 has been installed on BlueGene. To use Numpy and Scipy, the module essl/5.1 has to be loaded. The full python path has to be provided (otherwise the default version is used).
To use python on BlueGene (from within a job script or a debugjob session):
module load python/2.7.3 ##Only if you need numpy/scipy : module load xlf/14.1 essl/5.1 runjob --np 1 --ranks-per-node=1 --envs HOME=$HOME LD_LIBRARY_PATH=$LD_LIBRARY_PATH PYTHONPATH=/scinet/bgq/tools/Python/python2.7.3-20131205/lib/python2.7/site-packages/ : /scinet/bgq/tools/Python/python2.7.3-20131205/bin/python2.7 /PATHOFYOURSCRIPT.py
If you want to use the mmap python API, you must use it in PRIVATE mode as shown in the bellow example :
import mmap mm=mmap.mmap(-1,256,mmap.MAP_PRIVATE) mm.close()
Alternatively, you can use the mpi4py and h5py modules.
Also, please read Cython documentation.
Documentation
- BGQ Day: Introduction to Using the BG/Q Slides (updated in 2015) / Video recording (direct link)
- BGQ Day: BG/Q Hardware Overview Slides / Video recording (direct link)
- Julich BGQ Documentation
- Argonne Mira BGQ Wiki
- LLNL Sequoia BGQ Info
- Argonne MiraCon Presentations
- IBM Red Books BGQ System Administration Guide
- IBM Red Books BGQ Application Development
- IBM XL C/C++ for Blue Gene/Q: Getting started
- IBM XL C/C++ for Blue Gene/Q: Compiler reference
- IBM XL C/C++ for Blue Gene/Q: Language reference
- IBM XL C/C++ for Blue Gene/Q: Optimization and Programming Guide
- IBM XL Fortran for Blue Gene/Q: Getting started
- IBM XL Fortran for Blue Gene/Q: Compiler reference
- IBM XL Fortran for Blue Gene/Q: Language reference
- IBM XL Fortran for Blue Gene/Q: Optimization and Programming Guide
- IBM ESSL (Engineering and Scientific Subroutine Library) 5.1 for Linux on Power
- Allinea DDT 4.1 User Guide
- IBM LoadLeveler 5.1