Niagara Quickstart
| Niagara | |
|---|---|
| Installed | Jan 2018 |
| Operating System | CentOS 7.4 |
| Number of Nodes | 1500 nodes (60,000 cores) |
| Interconnect | Mellanox Dragonfly+ |
| Ram/Node | 188 GiB / 202 GB |
| Cores/Node | 40 (80 hyperthreads) |
| Login/Devel Node | niagara.scinet.utoronto.ca |
| Vendor Compilers | icc (C) ifort (fortran) icpc (C++) |
| Queue Submission | Slurm |
Specifications
The Niagara cluster is a large cluster of 1500 Lenovo SD350 servers each with 40 Intel "Skylake" cores at 2.4 GHz. The peak performance of the cluster is 3.02 PFlops delivered / 4.6 PFlops theoretical (would've been #42 on the TOP500 in Nov 2017). Each node has 188 GiB / 202 GB RAM per node (at least 4 GiB/core for user jobs. Being designed for large parallel workloads, it has a fast interconnect consisting of EDR InfiniBand in a Dragonfly+ topology with Adaptive Routing. The compute nodes are accessed through a queueing system that allows jobs with a minimum of 15 minutes and a maximum of 12 or 24 hours and favours large jobs.
Using Niagara: Logging in
As with all SciNet and CC (Compute Canada) compute systems, access to Niagara is via ssh (secure shell) only.
To access SciNet systems, first open a terminal window (e.g. MobaXTerm on Windows).
Then ssh into the Niagara login nodes with your CC credentials:
$ ssh -Y MYCCUSERNAME@niagara.scinet.utoronto.ca
or
$ ssh -Y MYCCUSERNAME@niagara.computecanada.ca
- The Niagara login nodes are where you develop, edit, compile, prepare and submit jobs.
- These login nodes are not part of the Niagara compute cluster, but have the same architecture, operating system, and software stack.
- The optional
-Yis needed to open windows from the Niagara command-line onto your local X server. - To run on Niagara's compute nodes, you must submit a batch job.
Migration to Niagara
Migration for Existing Users of the GPC
Niagara is replacing the General Purpose Cluster (GPC) and the Tightly Coupled Cluster (TCS) at SciNet. The TCS was decommissioned last fall, and GPC will be decommissioned very soon: the compute nodes of the GPC will be decommissioned on April 21, 2018, while the storage attached to the GPC will be decommissioned on May 9, 2018.
Active GPC Users got access to the new system, Niagara, on April 9, 2018.
Users' home and project folder were last copied over from the GPC to Niagara on April 5th, 2018, except for files whose name start with a period that were in their home directories (these files were never synced).
It is the user's responsibility to copy over data generated on the GPC after April 5th, 2018.
Data stored in scratch has also not been transfered automatically. Users are to clean up their scratch space on the GPC as much as possible (remember it's temporary data!). Then they can transfer what they need using datamover nodes.
To enable this transfer, there will be a short period during which you can have access to Niagara as well as to the GPC storage resources. This period will end on May 9, 2018.
To copy substantial amounts of data (i.e.,more than 10 GB), please use the datamovers of both the GPC (called gpc-logindm01 and gpc-logindm02) and the Niagara datamovers (called nia-dm1 and nia-dm2). For instance, to copy a directory abc from your GPC scratch to your Niagara scratch directory, you can do the following:
$ ssh CCUSERNAME@niagara.computecanada.ca $ ssh nia-dm1 $ scp -r SCINETUSERNAME@gpc-logindm01:\$SCRATCH/abc $SCRATCH/abc
For many of you, CCUSERNAME amd SCINETUSERNAME will be the same. Make sure you use the slash (\) before the first $SCRATCH; it cause the value of scratch on the remote node (i.e., here, gpc-logindm01) to be used. Note that the gpc-logindm01 will ask for your SciNet password.
You can also go the other way:
$ ssh SCINETUSERNAME@login.scinet.utoronto.ca $ ssh gpc-logindm01 $ scp -r $SCRATCH/abc CCUSERNAME@nia-dm1:\$SCRATCH/abc
Again, pay attention to the slash in front of the last occurrence of $SCRATCH.
If you are using rsync, we advice to refrain from using the -a flags, and if using cp, refrain from using the -a and -p flags.
For Non-GPC Users
Those of you new to SciNet, but with 2018 RAC allocations on Niagara, will have your accounts created and ready for you to login.
New, non-RAC users: we are still working out the procedure to get access. If you can't wait, for now, you can follow the old route of requesting a SciNet Consortium Account on the CCDB site.
Locating your directories
home and scratch
You have a home and scratch directory on the system, whose locations will be given by
$HOME=/home/g/groupname/myccusername
$SCRATCH=/scratch/g/groupname/myccusername
For example:
nia-login07:~$ pwd /home/s/scinet/rzon nia-login07:~$ cd $SCRATCH nia-login07:rzon$ pwd /scratch/s/scinet/rzon
project and archive
Users from groups with RAC storage allocation will also have a project and/or archive directory.
$PROJECT=/project/g/groupname/myccusername
$ARCHIVE=/archive/g/groupname/myccusername
NOTE: Currently archive space is available only via HPSS
IMPORTANT: Future-proof your scripts
Use the environment variables (HOME, SCRATCH, PROJECT, ARCHIVE) instead of the actual paths! The paths may change in the future.
Storage
| location | quota | block size | expiration time | backed up | on login | on compute |
|---|---|---|---|---|---|---|
| $HOME | 100 GB | 1 MB | yes | yes | read-only | |
| $SCRATCH | 25 TB | 16 MB | 2 months | no | yes | yes |
| $PROJECT | by group allocation | 16 MB | yes | yes | yes | |
| $ARCHIVE | by group allocation | dual-copy | no | no | ||
| $BBUFFER | ? | 1 MB | very short | no | ? | ? |
- Compute nodes do not have local storage.
- Archive space is on HPSS.
- Backup means a recent snapshot, not an achive of all data that ever was.
$BBUFFERstands for the Burst Buffer, a faster parallel storage tier for temporary data.
Moving data
Move amounts less than 10GB through the login nodes.
- Only Niagara login nodes visible from outside SciNet.
- Use scp or rsync to niagara.scinet.utoronto.ca or niagara.computecanada.ca (no difference).
- This will time out for amounts larger than about 10GB.
Move amounts larger than 10GB through the datamover nodes.
- From a Niagara login node, ssh to
nia-datamover1ornia-datamover2. - Transfers must originate from this datamover.
- The other side (e.g. your machine) must be reachable from the outside.
- If you do this often, consider using Globus, a web-based tool for data transfer.
Moving data to HPSS/Archive/Nearline using the scheduler.
- HPSS is a tape-based storage solution, and is SciNet's nearline a.k.a. archive facility.
- Storage space on HPSS is allocated through the annual Compute Canada RAC allocation.
Loading Software Modules
Other than essentials, all installed software is made available using module commands. These modules set environment variables (PATH, etc.) This allows multiple, conflicting versions of a given package to be available. module spider shows the available software.
For example:
nia-login07:~$ module spider
---------------------------------------------------
The following is a list of the modules currently av
---------------------------------------------------
CCEnv: CCEnv
NiaEnv: NiaEnv/2018a
anaconda2: anaconda2/5.1.0
anaconda3: anaconda3/5.1.0
autotools: autotools/2017
autoconf, automake, and libtool
boost: boost/1.66.0
cfitsio: cfitsio/3.430
cmake: cmake/3.10.2 cmake/3.10.3
...
Common module subcommands are:
module load <module-name>use particular software
module purgeremove currently loaded modules
module spider(or
module spider <module-name>)list available software packages
module availlist loadable software packages
module listlist loaded modules
On Niagara, there are really two software stacks:
A Niagara software stack tuned and compiled for this machine. This stack is available by default, but if not, can be reloaded with
module load NiaEnv
The same software stack available on Compute Canada's General Purpose clusters Graham and Cedar, compiled (for now) for a previous generation of CPUs:
module load CCEnv
If you want the same default modules loaded as on Cedar and Graham, then afterwards also
module load StdEnv.
Note: the *Env modules are sticky; remove them by --force.
Tips for loading software
We advise against loading modules in your .bashrc.
This could lead to very confusing behaviour under certain circumstances.
Instead, load modules by hand when needed, or by sourcing a separate script.
Load run-specific modules inside your job submission script.
Short names give default versions; e.g.
intel→intel/2018.2.It is usually better to be explicit about the versions, for future reproducibility.
Handy abbreviations:
ml → module list
ml NAME → module load NAME # if NAME is an existing module
ml X → module X
- Modules sometimes require other modules to be loaded first.
Solve these dependencies by using module spider.
Module spider
Oddly named, the module subcommand spider is the search-and-advice facility for modules.
Suppose one wanted to load the openmpi module. Upon trying to load the module, one may get the following message:
nia-login07:~$ module load openmpi Lmod has detected the error: These module(s) exist but cannot be loaded as requested: "openmpi" Try: "module spider openmpi" to see how to load the module(s).
So while that fails, following the advice that the command outputs, the next command would be:
nia-login07:~$ module spider openmpi
------------------------------------------------------------------------------------------------------
openmpi:
------------------------------------------------------------------------------------------------------
Versions:
openmpi/2.1.3
openmpi/3.0.1
openmpi/3.1.0rc3
------------------------------------------------------------------------------------------------------
For detailed information about a specific "openmpi" module (including how to load the modules) use
the module s full name.
For example:
$ module spider openmpi/3.1.0rc3
------------------------------------------------------------------------------------------------------
So this gives just more detailed suggestions on using the spider command. Following the advice again, one would type:
nia-login07:~$ module spider openmpi/3.1.0rc3
------------------------------------------------------------------------------------------------------
openmpi: openmpi/3.1.0rc3
------------------------------------------------------------------------------------------------------
You will need to load all module(s) on any one of the lines below before the "openmpi/3.1.0rc3"
module is available to load.
NiaEnv/2018a gcc/7.3.0
NiaEnv/2018a intel/2018.2
These are concrete instructions on how to load this particular openmpi module. Following these leads to a successful loading of the module.
nia-login07:~$ module load NiaEnv/2018a intel/2018.2 # note: NiaEnv is usually already loaded nia-login07:~$ module load openmpi/3.1.0rc3
nia-login07:~$ module list Currently Loaded Modules: 1) NiaEnv/2018a (S) 2) intel/2018.2 3) openmpi/3.1.0.rc3 Where: S: Module is Sticky, requires --force to unload or purge
Running Commercial Software
- Possibly, but you have to bring your own license for it.
- SciNet and Compute Canada have an extremely large and broad user base of thousands of users, so we cannot provide licenses for everyone's favorite software.
- Thus, the only commercial software installed on Niagara is software that can benefit everyone: Compilers, math libraries and debuggers.
- That means no Matlab, Gaussian, IDL,
- Open source alternatives like Octave, Python, R are available.
- We are happy to help you to install commercial software for which you have a license.
- In some cases, if you have a license, you can use software in the Compute Canada stack.
Compiling on Niagara: Example
Suppose one want to compile an application from two c source files, appl.c and module.c, which use the Gnu Scientific Library (GSL). This is an example of how this would be done:
nia-login07:~$ module list Currently Loaded Modules: 1) NiaEnv/2018a (S) Where: S: Module is Sticky, requires --force to unload or purge nia-login07:~$ module load intel/2018.2 gsl/2.4 nia-login07:~$ ls appl.c module.c nia-login07:~$ icc -c -O3 -xHost -o appl.o appl.c nia-login07:~$ icc -c -O3 -xHost -o module.o module.c nia-login07:~$ icc -o appl module.o appl.o -lgsl -mkl nia-login07:~$ ./appl
Note:
- The optimization flags -O3 -xHost allow the Intel compiler to use instructions specific to the architecture CPU that is present (instead of for more generic x86_64 CPUs).
- The GSL requires a cblas implementation, for is contained in the Intel Math Kernel Library (MKL). Linking with this library is easy when using the intel compiler, it just requires the -mkl flags.
- If compiling with gcc, the optimization flags would be -O3 -march=native. For the way to link with the MKL, it is suggested to use the MKL link line advisor.
Testing
You really should test your code before you submit it to the cluster to know if your code is correct and what kind of resources you need.
Small test jobs can be run on the login nodes.
Rule of thumb: couple of minutes, taking at most about 1-2GB of memory, couple of cores.
You can run the the ddt debugger on the login nodes after
module load ddt.Short tests that do not fit on a login node, or for which you need a dedicated node, request an
interactive debug job with the salloc commandnia-login07:~$ salloc -pdebug --nodes N --time=1:00:00
where N is the number of nodes. The duration of your interactive debug session can be at most one hour, can use at most 4 nodes, and each user can only have one such session at a time.
Alternatively, on Niagara, you can use the command
nia-login07:~$ debugjob N
where N is the number of nodes, If N=1, this gives an interactive session one 1 hour, when N=4 (the maximum), it give you 30 minutes.
Submitting jobs
Niagara uses SLURM as its job scheduler.
You submit jobs from a login node by passing a script to the sbatch command:
nia-login07:~$ sbatch jobscript.sh
This puts the job in the queue. It will run on the compute nodes in due course.
Jobs will run under their group's RRG allocation, or, if the group has none, under a RAS allocation (previously called `default' allocation).
Keep in mind:
Scheduling is by node, so in multiples of 40-cores.
Maximum walltime is 24 hours.
Jobs must write to your scratch or project directory (home is read-only on compute nodes).
Compute nodes have no internet access.
Download data you need beforehand on a login node.
Scheduling by Node
All job resource requests on Niagara are scheduled as a multiple of nodes.
- The nodes that your jobs run on are exclusively yours.
- No other users are running anything on them.
- You can ssh into them to see how things are going.
Whatever your requests to the scheduler, it will always be translated into a multiple of nodes.
Memory requests to the scheduler are of no use.
Your job gets N x 202GB of RAM if N is the number of nodes.
You should use all 40 cores on each of the nodes that your job uses.
You will be contacted if you don't, and we will help you get more science done.
Hyperthreading: Logical CPUs vs. cores
- Hyperthreading, a technology that leverages more of the physical hardware by pretending there are twice as many logical cores than real ones, is enabled on Niagara.
- So the OS and scheduler see 80 logical cores.
- 80 logical cores vs. 40 real cores typically gives about a 5-10% speedup (YMMV).
Because Niagara is scheduled by node, hyperthreading is actually fairly easy to use:
Ask for a certain number of nodes N for your jobs.
You know that you get 40xN cores, so you will use (at least) a total of 40xN mpi processes or threads.
(mpirun, srun, and the OS will automaticallly spread these over the real cores)
But you should also test if running 80xN mpi processes or threads gives you any speedup.
Regardless, your usage will be counted as 40xNx(walltime in years).
Example submission script (OpenMP)
#!/bin/bash #SBATCH --nodes=1 #SBATCH --cpus-per-task=40 #SBATCH --time=1:00:00 #SBATCH --job-name openmp_job #SBATCH --output=openmp_output_%j.txt cd $SLURM_SUBMIT_DIR module load intel/2018.2 export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK ./openmp_example # or "srun ./openmp_example".
Submit this script with the command:
nia-login07:~$ sbatch openmp_job.sh
- First line indicates that this is a bash script.
- Lines starting with
#SBATCHgo to SLURM. - sbatch reads these lines as a job request (which it gives the name
openmp_job) . - In this case, SLURM looks for one node with 40 cores to be run inside one task, for 1 hour.
- Once it found such a node, it runs the script:
- Change to the submission directory;
- Loads modules;
- Sets an environment variable;
- Runs the
openmp_exampleapplication.
- To use hyperthreading, just change
--cpus-per-task=40to--cpus-per-task=80.
Example submission script (MPI)
#!/bin/bash #SBATCH --nodes=8 #SBATCH --ntasks=320 #SBATCH --time=1:00:00 #SBATCH --job-name mpi_job #SBATCH --output=mpi_output_%j.txt cd $SLURM_SUBMIT_DIR module load intel/2018.2 module load openmpi/3.1.0rc3 mpirun ./mpi_example # or "srun ./mpi_example"
Submit this script with the command:
nia-login07:~$ sbatch mpi_job.sh
First line indicates that this is a bash script.
Lines starting with
#SBATCHgo to SLURM.sbatch reads these lines as a job request (which it gives the name
mpi_job)In this case, SLURM looks for 8 nodes with 40 cores on which to run 320 tasks, for 1 hour.
Once it found such a node, it runs the script:
- Change to the submission directory;
- Loads modules;
- Runs the
mpi_exampleapplication.
- To use hyperthreading, just change --ntasks=320 to --ntasks=640, and add --bind-to none to the mpirun command (the latter is necessary for OpenMPI only, not when using IntelMPI).
Monitoring queued jobs
Once the job is incorporated into the queue, there are some command you can use to monitor its progress.
squeueorqsumto show the job queue (squeue -u $USERfor just your jobs);squeue -j JOBIDto get information on a specific job(alternatively,
scontrol show job JOBID, which is more verbose).squeue --start -j JOBIDto get an estimate for when a job will run; these tend not to be very accurate predictions.scancel -i JOBIDto cancel the job.sinfo -pcomputeto look at available nodes.jobperf JOBIDto get an instantaneous view of the cpu and memory usage of the nodes of the job while it is running.sacctto get information on your recent jobs.More utilities like those that were available on the GPC are under development.
Data Management and I/O Tips
- $HOME, $SCRATCH, and $PROJECT all use the parallel file system called GPFS.
- Your files can be seen on all Niagara login and compute nodes.
- GPFS is a high-performance file system which provides rapid reads and writes to large data sets in parallel from many nodes.
- But accessing data sets which consist of many, small files leads to poor performance.
- Avoid reading and writing lots of small amounts of data to disk.
- Many small files on the system would waste space and would be slower to access, read and write.
- Write data out in binary. Faster and takes less space.
- The Burst Buffer is better for i/o heavy jobs and to speed up checkpoints.
Visualization
Software Available
We have installed the latest versions of the open source visualization suites: VMD, VisIt and ParaView.
Notice that for using ParaView you need to explicitly specify one of the mesa flags in order to avoid trying to use openGL, i.e., after loading the paraview module, use the following command:
paraview --mesa-swr
Notice that Niagara does not have specialized nodes nor specially designated hardware for visualization, so if you want to perform interactive visualization or exploration of your data you will need to submit an interactive job (debug job, see #Testing). For the same reason you won't be able to request or use GPUs for rendering as there are none!
Interactive Visualization
Runtime is limited on the login nodes, so you will need to request a testing job in order to have more time for exploring and visualizing your data. Additionally by doing so, you will have access to the 40 cores of each of the nodes requested. For performing an interactive visualization session in this way please follow these steps:
- ssh into niagara.scinet.utoronto.ca with the -X/-Y flag for x-forwarding
- request an interactive job, ie. debugjob this will connect you to a node, let's say for the argument "nia0xyz"
- run your favourite visualization program, eg. visit/paraview module load visit visit
- exit the debug session.
module load paraview paraview --mesa-swr |
Client-Server Mode
You can use any of the remote visualization protocols supported for both VisIt and ParaView.
Both, VisIt and ParaView, support "remote visualization" protocols. This includes:
- accessing data remotely, ie. stored on the cluster
- rendering visualizations using the compute nodes as rendering engines
- or both
VisIt Client-Server Configuration
For allowing VisIt connect to the Niagara cluster you need to set up a "Host Configuration".
Choose *one* of the methods bellow:
Niagara Host File Configuration
You can just download the Niagara host file, right click on the following link host_niagara.xml and select save as... Depending on the OS you are using on your local machine:
- on a Linux/Mac OS place this file in
~/.visit/hosts/ - on a Windows machine, place the file in
My Documents\VisIt 2.13.0\hosts\
Restart VisIt and check that the niagara profile should be available in your hosts.
Manual Niagara Host Configuration
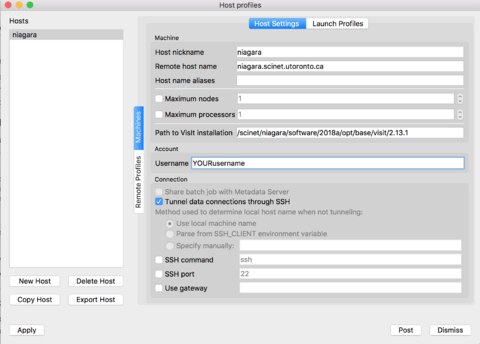
If you prefer to set up the verser yourself, instead of the configuration file from the previous section, just follow along these steps. Open VisIt in your computer, go to the 'Options' menu, and click on "Host profiles..." The click on 'New Host' and select:
Host nickname = niagara Remote host name = niagara.scinet.utoronto.ca Username = Enter_Your_OWN_username_HERE Path to VisIt installation = /scinet/niagara/software/2018a/opt/base/visit/2.13.1
Click on the "Tunnel data connections through SSH", and then hit Apply!
|
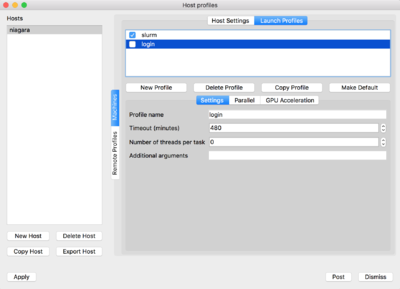
Now on the top of the window click on 'Launch Profiles' tab.
You will have to create two profiles:
-
login: for connecting through the login nodes and accessing data -
slurm: for using compute nodes as rendering engines
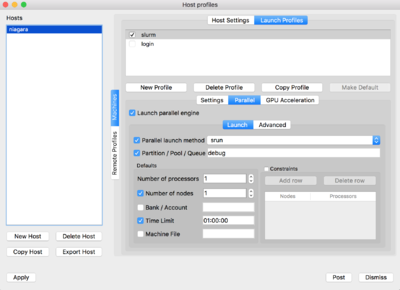
For doing so, click on 'New Profile', set the corresponding profile name, ie. login/slurm. Then click on the Parallel tab and set the "Launch parallel engine"
For the slurm profile, you will need to set the parameters as seen below:
|
|
Finally, after you are done with these changes, go to the "Options" menu and select "Save settings", so that your changes are saved and available next time you relaunch VisIt.
Final Considerations
Usually both VisIt and ParaView require to use the same version between the local client and the remote host, please try to sick to that to avoid having incompatibility issues, which might result in potential problems during the connections.
Other Versions
Alternatively you can try to use the visualization modules available on the CCEnv stack, for doing so just load the CCEnv module and select your favourite visualization module.
Further information
Useful sites
- SciNet: https://www.scinet.utoronto.ca
- Niagara: https://docs.computecanada.ca/wiki/niagara
- System Status: https://wiki.scinet.utoronto.ca/wiki/index.php/System_Alerts
- Training: https://support.scinet.utoronto.ca/education
Support
- support@scinet.utoronto.ca
- niagara@computecanada.ca
