SOSCIP GPU
| SOSCIP GPU | |
|---|---|
| Installed | September 2017 |
| Operating System | Ubuntu 16.04 Little-endian |
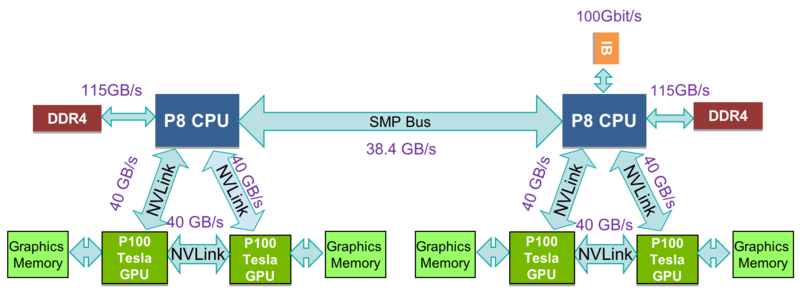
| Number of Nodes | 15x Power 8 with 4x NVIDIA P100 each node |
| Interconnect | Infiniband EDR |
| Ram/Node | 512 GB |
| Cores/Node | 2 x 10 core (20 physical, 160 SMT) |
| Login/Devel Node | sgc01 |
| Vendor Compilers | xlc/xlf, nvcc |
This is the wiki page for the soon-to-be-decommisioned SOSCIP GPU Cluster. The new GPU cluster, called Mist, replaces it. The instructions below will not work on Mist!
SOSCIP
The SOSCIP GPU Cluster is a Southern Ontario Smart Computing Innovation Platform (SOSCIP) resource located at the University of Toronto's SciNet HPC facility. The SOSCIP multi-university/industry consortium is funded by the Ontario Government and the Federal Economic Development Agency for Southern Ontario [1].
Support Email
Please use <soscip-support@scinet.utoronto.ca> for SOSCIP GPU specific inquiries.
Specifications
The SOSCIP GPU Cluster consists of of 15 (1 login/development + 14 compute) IBM Power 822LC "Minsky" Servers each with 2x10core 3.25GHz Power8 CPUs and 512GB Ram. Similar to Power 7, the Power 8 utilizes Simultaneous MultiThreading (SMT), but extends the design to 8 threads per core allowing the 20 physical cores to support up to 160 threads. Each node has 4x NVIDIA Tesla P100 GPUs each with 16GB of RAM with CUDA Capability 6.0 (Pascal) connected using NVlink.
Access and Login
In order to obtain access to the system, you must request access to the SOSCIP GPU Platform. Instructions will have been sent to your sponsoring faculty member via E-mail at the beginning of your SOSCIP project.
Access to the SOSCIP GPU Platform is provided through the BGQ login node, bgqdev.scinet.utoronto.ca using ssh, and from there you can proceed to the GPU development node sgc01-ib0 via ssh. Your user name and password is the same as it is for SciNet systems.
Filesystem
The filesystem is shared with the BGQ system. See here for details.
Job Submission
The SOSCIP GPU cluster uses SLURM as a job scheduler and jobs are scheduled by node, ie 20 cores and 4 GPUs each. Jobs are submitted from the login/development node sgc01. The maximum walltime per job is 12 hours (except in the 'long' queue, see Longer Jobs). The max number of nodes for each job is 4 (8 for 'long' queue).
- If a job cannot fully utilize all 4 GPUs in a node, please reference Packing single-GPU jobs
Job is submitted using sbatch command:
$ sbatch myjob.script
Where myjob.script is
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=20 # MPI tasks (needed for srun/mpirun) #SBATCH --time=1:00:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node cd $SLURM_SUBMIT_DIR hostname nvidia-smi
More information about the sbatch command is found here.
You can query job information using
squeue
You can list jobs sorted by priority, e.g. highest priority job at bottom:
squeue -S p
To see only your own jobs, run
squeue -u <userid>
Once your job is running, SLURM creates a file usually named slurm<jobid>.out in the directory from where you issued the sbatch command. This contains the console output from your job. You can monitor the output of your job by using the tail -f <file> command.
To cancel a job use
scancel $JOBID
Detailed job info
Default "squeue" out shows only basic information of the jobs. User can set SQUEUE_FORMAT environment variable to tell SLURM to genrate more detailed squeue output including estimated start time:
#It is recommended to put this command into user's ~/.bashrc file. export SQUEUE_FORMAT="%.8i %.8u %.12a %.10P %.20j %.3t %16S %.10L %.5D %.4C %.6b %.7m %N (%r) "
Longer jobs
If your job takes more than 12 hours or more than 4 nodes, the sbatch command will not let you submit your job. There is, however, a way to have jobs up to 24 hours long and up to 8 nodes, by specifying "-p long" as an option (i.e., add #SBATCH -p long to your job script). The priority of such jobs may be throttled in the future if we see that the 'long' queue is having a negative efffect on turnover time in the queue.
Interactive
For an interactive session use
salloc --gres=gpu:4 --time=1:00:00
After executing this command, you may have to wait in the queue until a system is available.
More information about the salloc command is here.
Automatic Resumption and Job Dependencies
Commonly you may have a job that you know will take longer to run than what is permissible in the queue. As long as your program contains checkpoint or restart capability, you can submit a series of Singleton jobs. Singleton dependency will execute jobs, which are having the same "job-name", one after another. User just needs to sbatch the same job script multiple times. More details of SLURM dependent job can be found in SBATCH_Help_Page. To submit singleton jobs, add these two lines in the job script:
#SBATCH --job-name=<same job name for a series of singleton jobs> #SBATCH --dependency=singleton
Packing single-GPU jobs within one SLURM job submission
Jobs are scheduled by node (4 GPUs) on SOSCIP GPU cluster. If user's code/program cannot utilize all 4 GPUs, user can use GNU Parallel tool to pack 4 or more single-GPU jobs into one SLURM job. Below is an example of submitting 4 single-GPU python codes within one job: (When using GNU parallel for a publication please cite as per parallel --citation)
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=20 # MPI tasks (needed for srun)
#SBATCH --time=00:10:00 # H:M:S
#SBATCH --gres=gpu:4 # Ask for 4 GPUs per node
. /etc/profile.d/modules.sh #enable module command
module load gnu-parallel/20180422
cd $SLURM_SUBMIT_DIR
parallel -a jobname-params.input --colsep ' ' -j 4 'CUDA_VISIBLE_DEVICES=$(( {%} - 1 )) numactl -N $(( ({%} -1) / 2 )) python {1} {2} {3} &> jobname-{#}.out'
The jobname-params.input file contains:
code-1.py --param1=a --param2=b code-2.py --param1=c --param2=d code-3.py --param1=e --param2=f code-4.py --param1=g --param2=h
- In the above example, GNU Parallel tool will read jobname-params.input file and separate parameters. Each row in the input file has to contain exact 3 parameters to python. code-N.py is also considered as a parameter. User can change parameter number in the parallel command ({1} {2} {3}...).
- "-j 4" flag limits the max number of jobs to be 4. User can have more rows in the input file, but GNU Parallel tool only executes maximum of 4 at the same time.
- "CUDA_VISIBLE_DEVICES=$(( {%} - 1 ))" will limit one GPU for each job. "numactl -N $(( ({%} -1) / 2 ))" will bind 2 jobs on CPU socket 0, other 2 jobs on socket 1. {%} is job slot which will be translated to 1 or 2 or 3 or 4 in this case.
- Outputs will be jobname-1.out, jobname-2.out,jobname-3.out,jobname-4.out... {#} is job number which will be translated to the row number in the input file.
- If you explicitly select GPU ID in your code, you need to always select 0.
- Do NOT change or set CUDA_VISIBLE_DEVICES environment variable in your code.
Packing two 2-GPU jobs
GPU 0 and 1 are connected with high speed NVLINK. GPU 2 and 3 are connected with NVLINK as well. There is no NVLINK between 0,1 and 2,3. User may want to run jobs with only 2 GPUs which are connected with NVLINK. To pack two 2-GPU jobs into one SLURM job, modify the above parallel tool command as below:
parallel -a jobname-params.input --colsep ' ' -j 2 'CUDA_VISIBLE_DEVICES=$(({%}*2-2)),$(({%}*2-1)) numactl -N $(({%} -1)) python {1} {2} {3} &> jobname-{#}.out'
This command will run 2 sub jobs at a time. The job 1 will use GPU id 0,1 and CPU socket 0. Job 2 will use GPU id 2,3 and CPU socket 1.
Compiler
CUDA
The current installed CUDA Tookits are 8.0, 9.0, 9.2 and 10.1.
module load cuda/<version>
The current NVIDIA driver version is 418.40.04
The driver is installed locally, however the CUDA Toolkit is installed in:
/scinet/sgc/Libraries/CUDA/8.0 /scinet/sgc/Libraries/CUDA/9.0 /scinet/sgc/Libraries/CUDA/9.2 /scinet/sgc/Libraries/CUDA/10.1
Documentation and API reference information for the CUDA Toolkit can be found here: http://docs.nvidia.com/cuda/index.html
GNU Compilers
System default compiler is GCC-5.4.0. More recent versions of the GNU Compiler Collection (C/C++/Fortran) are provided in the IBM Advance Toolchain with enhancements for the POWER8 CPU. To load the newer advance toolchain version use:
Advance Toolchain V11.0-3
module load gcc/7.3.1
More information about the IBM Advance Toolchain can be found here: https://developer.ibm.com/linuxonpower/advance-toolchain/
IBM XL Compilers
To load the native IBM xlc/xlc++ and xlf (Fortran) compilers, run
module load xlc/16.1.1 module load xlf/16.1.1
IBM XL Compilers are enabled for use with NVIDIA GPUs, including support for OpenMP 4.5 GPU offloading and integration with NVIDIA's nvcc command to compile host-side code for the POWER8 CPU.
Information about the IBM XL Compilers can be found at the following links:
OpenMPI
Currently OpenMPI has been setup on the 14 compute nodes connected over EDR Infiniband. Available modules are:
openmpi/4.0.1-gcc-5.4.0 openmpi/4.0.1-gcc-7.3.1 openmpi/4.0.1-xl-16.1.1 openmpi/3.1.4-gcc-5.4.0 openmpi/3.1.4-gcc-7.3.1 openmpi/3.1.4-xl-16.1.1 ibm-mpi/10.3 (IBM Spectrum MPI)
PGI
To load PGI compiler and its OpenMPI environment, run:
module load pgi/19.4 module load openmpi/3.1.3-pgi-19.4 OR module load pgi/18.10 module load openmpi/2.1.2-pgi-18.10
Software
Anaconda (Python)
Anaconda is a popular distribution of the Python programming language. It contains several common Python libraries such as SciPy and NumPy as pre-built packages, which eases installation. Anaconda is provided as modules: anaconda2 and anaconda3
To install Anaconda locally, user need to load the module and create a conda environment: (anaconda3 as example)
module load anaconda3 conda create -n myPythonEnv python=3.6
- Note: By default, conda environments are located in $HOME/.conda/envs. Downloaded tarballs and packages are under $SCRATCH/.conda/pkgs. $SCRATCH is not backed up.
To activate the conda environment: (should be activated before running python)
source activate myPythonEnv
Once the environment is activated, user can update or install packages via conda or pip
conda install -n myPythonEnv <package_name> (preferred way to install packages) pip install <package_name>
To deactivate:
source deactivate
To remove a conda enviroment:
conda remove --name myPythonEnv --all
To verify that the environment was removed, run:
conda info --envs
Bazel
Bazel is provided as modules on the system:
module load bazel/0.26.1
cuDNN
The NVIDIA CUDA Deep Neural Network library (cuDNN) is a GPU-accelerated library of primitives for deep neural networks. cuDNN accelerates widely used deep learning frameworks, including Caffe2, MATLAB, Microsoft Cognitive Toolkit, TensorFlow, Theano, and PyTorch. If a specific version of cuDNN is needed, user can download from https://developer.nvidia.com/cudnn and choose "cuDNN [VERSION] Library for Linux (Power8/Power9)". cuDNN versions are installed as modules:
cudnn/cuda9.2/7.5.0 cudnn/cuda10.1/7.6.5
CuPy
CuPy is an open-source matrix library accelerated with NVIDIA CUDA. It also uses CUDA-related libraries including cuBLAS, cuDNN, cuRand, cuSolver, cuSPARSE, cuFFT and NCCL to make full use of the GPU architecture. CuPy is an implementation of NumPy-compatible multi-dimensional array on CUDA. CuPy consists of the core multi-dimensional array class, cupy.ndarray, and many functions on it. It supports a subset of numpy.ndarray interface.
CuPy can be install into any conda environment. Python packages: numpy six and fastrlock are required. cuDNN and NCCL are optional.
module load cuda/10.1 cudnn/cuda10.1/7.5.1 nccl/cuda10.1/2.4.7 anaconda3 conda create -n cupy-env python=3.6 numpy six fastrlock source activate cupy-env CFLAGS="-I/scinet/sgc/Libraries/cuDNN/cuda10.1/7.5.1/include -I/scinet/sgc/Libraries/NCCL/cuda10.1/2.4.7/include" LDFLAGS="-L/scinet/sgc/Libraries/cuDNN/cuda10.1/7.5.1/lib -L/scinet/sgc/Libraries/NCCL/cuda10.1/2.4.7/lib" CUDA_PATH="/scinet/sgc/Libraries/CUDA/10.1" pip install cupy
GROMACS
GROMACS is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. It is primarily designed for biochemical molecules like proteins, lipids and nucleic acids that have a lot of complicated bonded interactions, but since GROMACS is extremely fast at calculating the nonbonded interactions (that usually dominate simulations) many groups are also using it for research on non-biological systems, e.g. polymers.
Available modules are: (please use "module show <module_name>" command to check pre-requested modules)
gromacs/2016.5-gcc5-ompi312 gromacs/2018.6-gcc7-ompi313 gromacs/2019.1-gcc7-ompi313
A two-node job example is below:
#!/bin/sh #SBATCH --nodes=2 #SBATCH --ntasks-per-node=20 # MPI tasks (needed for mpirun/srun) #SBATCH --time=1:00:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node . /etc/profile.d/modules.sh #enable module command module purge module load cuda/9.2 gcc/7.3.1 openmpi/3.1.3-gcc-7.3.1 gromacs/2018.6-gcc7-ompi313 cd $SLURM_SUBMIT_DIR mpirun gmx_mpi mdrun -ntomp 2 -pin on -s nvt_1us.tpr # 2 SMT (Simultaneous MultiThreading) per CPU core is recommended. There are 20 CPU cores per node, with SMT=2, there are 40 threads per node. # User is suggested to benchmark with different number of MPI ranks and number of threads per rank. Here we use 20 ranks per node, 2 threads per rank. # "-pin on" flag should always be used.
Horovod
Horovod is a distributed training framework for TensorFlow, Keras, and PyTorch. The goal of Horovod is to make distributed Deep Learning fast and easy to use.
To use Horovod on SOSCIP GPU cluster, user should have TensorFlow or PyTorch installed first then load the modules: (plus anaconda2/3 and cudnn modules for DL frameworks)
module load openmpi/4.0.1-gcc-5.4.0 cuda/9.2 nccl/2.4.2
Horovod can be installed by pip (in a Conda environment or system python's virtual environment) with the following configuration:
conda install cffi pycparser cloudpickle psutil HOROVOD_CUDA_HOME=/scinet/sgc/Libraries/CUDA/9.2 HOROVOD_NCCL_HOME=/scinet/sgc/Libraries/NCCL/cuda9.2/2.4.2/ HOROVOD_GPU_ALLREDUCE=NCCL pip install --no-cache-dir horovod
A multi-node Tensorflow-benchmarks example is below:
git clone https://github.com/tensorflow/benchmarks.git (and checkout branch for certain version of TensorFlow)
A 2-node job script:
#!/bin/bash #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 # MPI tasks (needed for srun/mpirun) #SBATCH --cpus-per-task=40 # One physical core shows as 8 logical cores #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node module load openmpi/4.0.1-gcc-5.4.0 cuda/9.2 nccl/2.4.2 #adding anaconda2/3 and cudnn modules for TensorFlow export OMP_NUM_THREADS=1 #Anaconda's numpy package on ppc64le with OpenBLAS multithreading may lead to incorrect answers #User also needs to setup TensorFlow environment as well mpirun -bind-to core -map-by slot:PE=5 -report-bindings -x NCCL_DEBUG=INFO -x LD_LIBRARY_PATH -x PATH -x OMP_NUM_THREADS -x HOROVOD_MPI_THREADS_DISABLE=1 python -u scripts/tf_cnn_benchmarks/tf_cnn_benchmarks.py --model resnet50 --batch_size 32 --variable_update horovod
4 tasks is required per node. This will create 4 MPI ranks per node, 1 rank per GPU. For each rank, it has 5 slots (-bind-to core -map-by slot:PE=5), which are 5 physical cores (in total 40 threads). User will see the detail binding information with -report-bindings flag.
Scaling tests, results in images/sec: (ResNet-50 synthetic data, batch size 32 per GPU)
1 GPU: 220.08 2 GPUs: 432.99 4 GPUs: 851.12 8 GPUs: 1676.43 16 GPUs: 3346.44 32 GPUs: 6635.06 60 GPUs: 12266.48
IBM PowerAI (v1.6.0 or newer)
The PowerAI ML/DL packages are distributed as Conda packages in an online Conda repository. Load Anaconda3 module and use the PowerAI channel to install packages:
module load anaconda3 conda create --name powerai -c https://public.dhe.ibm.com/ibmdl/export/pub/software/server/ibm-ai/conda/ python=3.6 (or 3.7) source activate powerai
All the ML/DL frameworks can be installed at the same time by using the powerai meta-package. With the Conda environment you want to install in activated run:
conda install -c https://public.dhe.ibm.com/ibmdl/export/pub/software/server/ibm-ai/conda/ python=3.6(or 3.7) <package name>
Available packages:
caffe ddl pytorch tensorflow-gpu pai4sk snapml-spark cudf cuml
Keras
Keras is a popular high-level deep learning software development framework. It runs on top of other deep-learning frameworks such as TensorFlow.
- The easiest way to install Keras is to install Anaconda first, then install Keras by using using the pip install (not conda install) command in a Conda environment. Keras uses TensorFlow underneath to run neural network models. Before running code using Keras, be sure to install TensorFlow wheel.
NAMD
NAMD is a parallel, object-oriented molecular dynamics code designed for high-performance simulation of large biomolecular systems. NAMD is installed on SOSCIP GPU cluster as modules. If a new version is required or if for some reason you need to do your own installation, please contact sosicp-support email.
v2.13 (for single or multi-node via PAMI and IBM Spectrum MPI)
2.13 versions of NAMD supports multi-node simulations via PAMI. Available modules are:
namd/2.13-gcc7-pami namd/2.13-xl16-pami
An example of the job script (using 2 nodes, one process per node, 40 CPU threads per process + 4 GPUs per process):
#!/bin/bash #SBATCH --nodes=2 #SBATCH --ntasks-per-node=1 # MPI tasks (needed for mpirun/srun) #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node . /etc/profile.d/modules.sh #enable module command module purge module load gcc/7.3.1 cuda/9.2 powerAI-5.3/mldl-spectrum namd/2.13-gcc7-pami cd $SLURM_SUBMIT_DIR scontrol show hostnames > slurm-nodelist `which charmrun` -npernode $SLURM_NTASKS_PER_NODE -hostfile slurm-nodelist `which namd2` +setcpuaffinity +pemap 0-159:4 +idlepoll +ppn 40 +p $((SLURM_NTASKS*40)) stmv.namd
The above example runs one process per node. NAMD may scale better if using one process per GPU device. User is recommended to benchmark with both configurations. A one process per GPU device example is below:
#!/bin/bash #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 # MPI tasks (needed for mpirun/srun) #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node . /etc/profile.d/modules.sh #enable module command module purge module load gcc/7.3.1 cuda/9.2 powerAI-5.3/mldl-spectrum namd/2.13-gcc7-pami cd $SLURM_SUBMIT_DIR scontrol show hostnames > slurm-nodelist `which charmrun` -npernode $SLURM_NTASKS_PER_NODE -hostfile slurm-nodelist `which namd2` +setcpuaffinity +pemap 0-159:4 +idlepoll +ppn 10 +p $((SLURM_NTASKS*10)) stmv.namd
NCCL
The NVIDIA Collective Communications Library (NCCL) implements multi-GPU and multi-node collective communication primitives that are performance optimized for NVIDIA GPUs. NCCL is provided as modules on the system:
nccl/2.4.2 nccl/cuda10.1/2.4.7
PyTorch
The PyTorch which is included in PowerAI or Anaconda may not be the most recent version. Newer PyTorch is provided as prebuilt Python Wheel that users can use pip to install under user space. Custom Python wheels are stored in /scinet/sgc/Applications/PyTorch_wheels/conda. It is highly recommended to install custom PyTorch wheels into a Conda environment.
Installing with Anaconda2 (Python2.7):
- Load modules:
module load cuda/9.2 cudnn/cuda9.2/7.4.1 anaconda2
- Create a conda environment pytorch-1.1.0a0+87ae155-py2:
conda create -n pytorch-1.1.0a0+87ae155-py2 python=2.7
- Activate conda environment:
source activate pytorch-1.1.0a0+87ae155-py2
- Install PyTorch into the conda environment with updated dependencies:
conda install -n pytorch-1.1.0a0+87ae155-py2 python-lmdb python-leveldb numpy pyyaml cmake cffi typing cython snappy future pip install /scinet/sgc/Libraries/protobuf/3.6.1/python/protobuf-3.6.1-cp27-cp27mu-linux_ppc64le.whl pip install /scinet/sgc/Applications/PyTorch_wheels/conda/torch-1.1.0a0+87ae155-cp27-cp27mu-linux_ppc64le.whl
Installing with Anaconda3 (Python3.6):
- Load modules:
module load cuda/9.2 cudnn/cuda9.2/7.4.1 anaconda3
- Create a conda environment pytorch-1.1.0a0+87ae155-py3:
conda create -n pytorch-1.1.0a0+87ae155-py3 python=3.6
- Activate conda environment:
source activate pytorch-1.1.0a0+87ae155-py3
- Install PyTorch into the conda environment with updated dependencies:
conda install -n pytorch-1.1.0a0+87ae155-py3 python-lmdb python-leveldb numpy pyyaml cmake cffi typing cython snappy future pydot pip install /scinet/sgc/Libraries/protobuf/3.6.1/python/protobuf-3.6.1-cp36-cp36m-linux_ppc64le.whl pip install /scinet/sgc/Applications/PyTorch_wheels/conda/torch-1.1.0a0+87ae155-cp36-cp36m-linux_ppc64le.whl
Submitting jobs
The above myjob.script file needs to be modified to run custom PyTorch. Required modules need to be loaded. Conda environment needs to be activated.
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=20 # MPI tasks (needed for srun/mpirun) #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node . /etc/profile.d/modules.sh #enable module command module purge module load cuda/9.2 cudnn/cuda9.2/7.4.1 anaconda3 source activate pytorch-1.1.0a0+87ae155-py3 export OMP_NUM_THREADS=1 #Anaconda's numpy package on ppc64le with OpenBLAS multithreading may lead to incorrect answers cd $SLURM_SUBMIT_DIR python code.py
RAPIDS
The RAPIDS is a suite of open source software libraries that gives you the freedom to execute end-to-end data science and analytics pipelines entirely on GPUs. The RAPIDS data science framework includes a collection of libraries: cuDF(GPU DataFrames), cuML(GPU Machine Learning Algorithms), cuStrings(GPU String Manipulation), etc.
To use PAPIDS on SOSCIP GPU platform, user need to load modules:
module load cuda/9.2 anaconda3 apache-arrow/0.12.1 custrings/0.3.0 cudf/0.5.1 cuml/0.5.1
Create conda environment without dependencies installed by conda and pip (after activate conda environment):
conda create -n rapids python=3.6 numba pandas cffi distributed cython pytest sphinx sphinxcontrib-websupport sphinx_rtd_theme numpydoc pandoc requests networkx
source activate rapids
pip install sphinx-markdown-tables nbsphinx recommonmark
Install python bindings into the conda environment using pip:
pip install /scinet/sgc/Libraries/apache-arrow/0.12.1/python/pyarrow-0.12.1a0-cp36-cp36m-linux_ppc64le.whl pip install /scinet/sgc/Libraries/custrings/0.3.0/python/nvstrings_cudaunknown-0.0.0.dev0-cp36-cp36m-linux_ppc64le.whl pip install /scinet/sgc/Libraries/cudf/0.5.1/python/cudf-0.6.0.dev0+1714.gabea679.dirty-cp36-cp36m-linux_ppc64le.whl pip install /scinet/sgc/Libraries/cudf/0.5.1/python/libgdf_cffi-0.6.0-py3-none-any.whl pip install /scinet/sgc/Libraries/cudf/0.5.1/python/librmm_cffi-0.5.0-py3-none-any.whl pip install /scinet/sgc/Libraries/cuml/0.5.1/python/cuml-0.5.1+711.g21b15ea.dirty-cp36-cp36m-linux_ppc64le.whl (optional) pip install /scinet/sgc/Libraries/scipy/scipy-1.2.1-cp36-cp36m-linux_ppc64le.whl (optional) pip install /scinet/sgc/Libraries/scikit-learn/scikit_learn-0.20.3-cp36-cp36m-linux_ppc64le.whl
If newer version of RAPIDS is needed, please contact <soscip-support@scinet.utoronto.ca>
TensorFlow (v2.0)
The TensorFlow which is included in PowerAI or Anaconda may not be the most recent version. Newer versions of TensorFlow are provided as prebuilt Python Wheels that users can use pip to install under user space. Python wheels are stored in /scinet/sgc/Applications/TensorFlow_wheels/conda. It is highly recommended to install TensorFlow wheels into a Conda virtual environment.
- Starting from v1.12, TensorFlow is built with XLA support by default. The TensorFlow/XLA JIT compiler compiles and runs parts of TensorFlow graphs via XLA. The benefit of this over the standard TensorFlow implementation is that XLA can fuse multiple operators (kernel fusion) into a small number of compiled kernels. Fusing operators can reduce memory bandwidth requirements and improve performance compared to executing operators one-at-a-time, as the TensorFlow executor does. To use XLA, please check Using JIT for instructions.
Installing with Anaconda3 (Python3.7):
- Load modules:
module load cuda/10.1 cudnn/cuda10.1/7.6.4 tensorrt/6.0.1.5 anaconda3
- Create a conda environment tensorflow-2.0-py3:
conda create -n tensorflow-2.0-py3 python=3.7
- Activate conda environment:
source activate tensorflow-2.0-py3
- Install TensorFlow into the conda environment with updated dependencies:
conda install -n tensorflow-2.0-py3 conda install keras-applications keras-preprocessing 'future>=0.17.1' mock cython numpy protobuf grpcio markdown html5lib werkzeug absl-py bleach six h5py astor gast==0.2.2 termcolor setuptools wrapt google-pasta
pip install /scinet/sgc/Applications/TensorFlow_wheels/conda/tensorflow-2.0.0-cp37-cp37m-linux_ppc64le.whl
Submitting jobs
The above myjob.script file needs to be modified to run custom TensorFlow. Required modules need to be loaded. Conda environment needs to be activated.
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=20 # MPI tasks (needed for srun/mpirun) #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node . /etc/profile.d/modules.sh #enable module command module purge module load cuda/10.1 cudnn/cuda10.1/7.6.4 tensorrt/6.0.1.5 anaconda3 source activate tensorflow-2.0-py3 export OMP_NUM_THREADS=1 #Anaconda's numpy package on ppc64le with OpenBLAS multithreading may lead to incorrect answers cd $SLURM_SUBMIT_DIR python code.py
TensorFlow Probability
TensorFlow Probability is a Python library built on TensorFlow that makes it easy to combine probabilistic models and deep learning on modern hardware (TPU, GPU). User needs to install TensorFlow in a conda environment first, then uses pip to install TensorFlow Probability:
pip install --upgrade tensorflow-probability
VASP
The Vienna Ab initio Simulation Package (VASP) is a computer program for atomic scale materials modelling, e.g. electronic structure calculations and quantum-mechanical molecular dynamics, from first principles. SOSCIP doesn't provide any VASP copy. User needs to obtain VASP source code after purchase and get permission to build/install the GPU version of VASP on SOSCIP GPU cluster. Please contact <soscip-support@scinet.utoronto.ca> if help is needed.
A makefile.include example (for VASP 5.4.4 + patch.5.4.4.16052018) is provided below: (User needs to load cuda/9.2 gcc/7.3.1 openmpi/4.0.1-gcc-7.3.1 modules before building VASP.)
# Precompiler options
CPP_OPTIONS= -DHOST=\"LinuxGNU\" \
-DMPI -DMPI_BLOCK=8000 \
-Duse_collective \
-DscaLAPACK \
-DCACHE_SIZE=4000 \
-Davoidalloc \
-Duse_bse_te \
-Dtbdyn \
-Duse_shmem
CPP = gcc -mcpu=power8 -E -P -C -w $*$(FUFFIX) >$*$(SUFFIX) $(CPP_OPTIONS)
FC = mpif90 -mcpu=power8
FCL = mpif90 -mcpu=power8
FREE = -ffree-form -ffree-line-length-none
FFLAGS = -w
OFLAG = -O2
OFLAG_IN = $(OFLAG)
DEBUG = -O0
LIBDIR =
BLAS = -fopenmp /scinet/sgc/Applications/PowerAI/5.3/openblas/lib/libopenblas_power8p-r0.3.2.a
LAPACK = -fopenmp /scinet/sgc/Applications/PowerAI/5.3/openblas/lib/libopenblas_power8p-r0.3.2.a
BLACS =
SCALAPACK = /scinet/sgc/Libraries/scalapack/2.0.2/libscalapack.a
LLIBS = $(SCALAPACK) $(LAPACK) $(BLAS)
FFTW ?= /scinet/sgc/Libraries/fftw/3.3.8
LLIBS += /scinet/sgc/Libraries/fftw/3.3.8/lib/libfftw3.a
INCS = -I$(FFTW)/include
OBJECTS = fftmpiw.o fftmpi_map.o fftw3d.o fft3dlib.o \
/scinet/sgc/Libraries/fftw/3.3.8/lib/libfftw3.a
OBJECTS_O1 += fftw3d.o fftmpi.o fftmpiw.o
OBJECTS_O2 += fft3dlib.o
# For what used to be vasp.5.lib
CPP_LIB = $(CPP)
FC_LIB = $(FC)
CC_LIB = gcc -mcpu=power8
CFLAGS_LIB = -O
FFLAGS_LIB = -O1
FREE_LIB = $(FREE)
OBJECTS_LIB= linpack_double.o getshmem.o
# For the parser library
CXX_PARS = g++ -mcpu=power8
LIBS += parser
LLIBS += -Lparser -lparser -lstdc++
# Normally no need to change this
SRCDIR = ../../src
BINDIR = ../../bin
#================================================
# GPU Stuff
CPP_GPU = -DCUDA_GPU -DRPROMU_CPROJ_OVERLAP -DCUFFT_MIN=28 -UscaLAPACK #-DUSE_PINNED_MEMORY
OBJECTS_GPU = fftmpiw.o fftmpi_map.o fft3dlib.o fftw3d_gpu.o fftmpiw_gpu.o
CC = gcc -mcpu=power8
CXX = g++ -mcpu=power8
CFLAGS = -fPIC -fopenmp -DMAGMA_WITH_MKL -DMAGMA_SETAFFINITY -DADD_ -DGPUSHMEM=300 -DHAVE_CUBLAS
CUDA_ROOT ?= /usr/local/cuda-9.2
NVCC := $(CUDA_ROOT)/bin/nvcc -Xcompiler -U__FLOAT128__
CUDA_LIB := -L$(CUDA_ROOT)/lib64 -lnvToolsExt -lcudart -lcuda -lcufft -lcublas
GENCODE_ARCH := -gencode=arch=compute_60,code=\"sm_60,compute_60\"
MPI_INC = /scinet/sgc/mpi/openmpi/4.0.1-gcc7.3.1-mlxofed-knem/include
An example job script is below:
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=4 # MPI tasks (needed for srun/mpirun) #SBATCH --time=00:10:00 # H:M:S #SBATCH --gres=gpu:4 # Ask for 4 GPUs per node module purge module load cuda/9.2 gcc/7.3.1 openmpi/4.0.1-gcc-7.3.1 mpirun -bind-to core -npersocket $((SLURM_NTASKS_PER_NODE/2)) -report-bindings -x OMP_NUM_THREADS=1 /path/to/vasp/vasp.5.4.4/bin/vasp_gpu #OMP_NUM_THREADS=1 has to be set. This example runs 1 MPI rank per GPU. User is suggested to benchmark with 2 or more MPI ranks per GPU. Nvidia Multi-Process Service is also suggested to be tested when using multiple MPI ranks per GPU.
Performance Guide
CPU Performance
Simultaneous multithreading (SMT)
POWER8 is designed to be a massively multithreaded chip, with each of its cores capable of handling 8 hardware threads simultaneously, for a total of 160 threads executed simultaneously on SOSCIP GPU node with 20 physical cores. On the system, it will show 160 (logical) CPU cores: CPU 0-7 is physical core 0, CPU 8-15 is physical core 1, ... , CPU 152-159 is physical core 19. Many of the programs developed on Intel/AMD x86 system are not optimized for POWER8 CPU. Using up all 8 hardware threads may significantly slow down the performance. Many programs show best performance with only 1 or 2 threads per physical core.
A common problem is thread binding. Software like GROMACS and NAMD can automatically bind certain number of threads to physical cores. If setting 2 threads per physical core, Gromacs/NAMD will use CPU 0,4,8,12,16, ..., 152, 156 only. Many Deep Learning softwares including TensorFlow and Pytorch are not able to automatically bind threads to a certain core. In this case, user can manually force the program to use certain CPUs via numactl tool.
If using 1 thread each physical core: numactl -C 0,8,16,24,32,40,48,56,64,72,80,88,96,104,112,120,128,136,144,152 python code.py If using 2 threads each physical core: numactl -C 0,4,8,12,16,20,24,28,32,36,40,44,48,52,56,60,64,68,72,76,80,84,88,92,96,100,104,108,112,116,120,124,128,132,136,140,144,148,152,156 python code.py
- MPI program cannot easily use numactl for thread binding. A rank file is used to bind rank to specific hardware thread(s). An example of a rank file, which uses 2 hardware threads per physical core, is shown below:
rank 0=sgc01 slot=0,4,8,12,16,20,24,28,32,36 rank 1=sgc01 slot=40,44,48,52,56,60,64,68,72,76 rank 2=sgc01 slot=80,84,88,92,96,100,104,108,112,116 rank 3=sgc01 slot=120,124,128,132,136,140,144,148,152,156
MPI program needs to be launched with flags: "-bind-to hwthread -rf myrankfile".
A rank file can be created automatically based on the node list generated by SLURM command "scontrol show hostnames". An example of python code to generate rankfile based on slurm nodelist:
#This python code generates rankfile with 4 MPI ranks per node, 5 physical cores per rank, 1 hardware thread per physical core
with open("slurm-nodelist", "r") as ins:
i=0
for line in ins:
print('rank '+str(i)+'='+line[:-1]+' slot=0,8,16,24,32')
print('rank '+str(i+1)+'='+line[:-1]+' slot=40,48,56,64,72')
print('rank '+str(i+2)+'='+line[:-1]+' slot=80,88,96,104,112')
print('rank '+str(i+3)+'='+line[:-1]+' slot=120,128,136,144,152')
i+=4
Job example:
scontrol show hostnames > slurm-nodelist python ~/createrankfile.py > myrankfile mpirun -bind-to hwthread -rf myrankfile -report-bindings yourprogram ...
GPU Performance
Profiling with NVVP
The NVIDIA Visual Profiler (NVVP) is a cross-platform performance profiling tool that delivers developers vital feedback for optimizing CUDA C/C++ applications. To use NVVP, user needs to enable X11 forwarding when ssh to sgc01: (Mac OS user needs to install XQuart)
ssh -Y bgqdev.scinet.utoronto.ca ssh -Y sgc01-ib0
CUDA module is required to use NVVP:
module load cuda/9.2 nvvp
Developing with Nsight
NVIDIA Nsight™ Eclipse Edition is a full-featured IDE powered by the Eclipse platform that provides an all-in-one integrated environment to edit, build, debug and profile CUDA-C applications. X11 forwarding is also required to use Nsight. After login with X11 enabled:
module load cuda/9.2 nsight
I/O Performance
GPFS is a high-performance filesystem which provides rapid reads and writes to large datasets in parallel from many nodes. As a consequence of this design, however, the file system performs quite poorly at accessing data sets which consist of many, small files. If user has to process a large number of small files, local SSD or RAM-disk can be used. Please contact <soscip-support@scinet.utoronto.ca> for more information.
NVLINK/Interconnection/Network Performance
Job Monitoring
GPU Usage Report from NVIDIA Management Library (NVML)
A per-process GPU utilization report is generated after each job execution. The report is placed at the end of the job output file, i.e. slurm-<jobid>.out.
- According to NVIDIA, enabling accounting mode has no negative impact on the GPU performance. See NVML_API_Reference_Guide
- An example output:
*********GPU usage report: (empty if GPU is not used)********* ===== Node sgc13 ===== sgc13: === GPU 0 === sgc13: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc13: 122244, 589355 ms, 64 %, 2 %, 297 MiB sgc13: 122248, 587600 ms, 65 %, 2 %, 297 MiB sgc13: 122246, 587603 ms, 65 %, 2 %, 299 MiB sgc13: 122245, 587611 ms, 65 %, 2 %, 297 MiB sgc13: 122247, 587620 ms, 65 %, 2 %, 299 MiB sgc13: 1526, 589370 ms, 64 %, 2 %, 0 MiB sgc13: === GPU 1 === sgc13: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc13: 122252, 587512 ms, 68 %, 2 %, 299 MiB sgc13: 122244, 589350 ms, 66 %, 2 %, 287 MiB sgc13: 122250, 587525 ms, 68 %, 2 %, 297 MiB sgc13: 122251, 587539 ms, 68 %, 2 %, 299 MiB sgc13: 122253, 587531 ms, 68 %, 2 %, 297 MiB sgc13: 122249, 587543 ms, 68 %, 2 %, 297 MiB sgc13: 1526, 589368 ms, 66 %, 2 %, 0 MiB sgc13: === GPU 2 === sgc13: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc13: 122257, 587428 ms, 64 %, 2 %, 299 MiB sgc13: 122244, 589345 ms, 63 %, 2 %, 287 MiB sgc13: 122255, 587445 ms, 64 %, 2 %, 297 MiB sgc13: 122256, 587438 ms, 64 %, 2 %, 299 MiB sgc13: 122258, 587451 ms, 64 %, 2 %, 297 MiB sgc13: 122254, 587452 ms, 64 %, 2 %, 297 MiB sgc13: 1526, 589364 ms, 63 %, 2 %, 0 MiB sgc13: === GPU 3 === sgc13: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc13: 122259, 587266 ms, 66 %, 2 %, 297 MiB sgc13: 122260, 587344 ms, 66 %, 2 %, 297 MiB sgc13: 122244, 589341 ms, 66 %, 2 %, 287 MiB sgc13: 122263, 587353 ms, 66 %, 2 %, 297 MiB sgc13: 122261, 587358 ms, 66 %, 2 %, 299 MiB sgc13: 122262, 587372 ms, 66 %, 2 %, 299 MiB sgc13: 1526, 589360 ms, 66 %, 2 %, 0 MiB ===== Node sgc15 ===== sgc15: === GPU 0 === sgc15: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc15: 38671, 587503 ms, 65 %, 2 %, 297 MiB sgc15: 38668, 587582 ms, 65 %, 2 %, 297 MiB sgc15: 38669, 587596 ms, 65 %, 2 %, 299 MiB sgc15: 38667, 588223 ms, 64 %, 2 %, 297 MiB sgc15: 38670, 587606 ms, 65 %, 2 %, 299 MiB sgc15: 1583, 588255 ms, 64 %, 2 %, 0 MiB sgc15: === GPU 1 === sgc15: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc15: 38672, 587502 ms, 66 %, 2 %, 297 MiB sgc15: 38674, 587514 ms, 66 %, 2 %, 299 MiB sgc15: 38675, 587517 ms, 66 %, 2 %, 299 MiB sgc15: 38667, 588219 ms, 66 %, 2 %, 287 MiB sgc15: 38673, 587520 ms, 66 %, 2 %, 297 MiB sgc15: 38676, 587528 ms, 66 %, 2 %, 297 MiB sgc15: 1583, 588254 ms, 66 %, 2 %, 0 MiB sgc15: === GPU 2 === sgc15: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc15: 38679, 587440 ms, 66 %, 2 %, 299 MiB sgc15: 38667, 588214 ms, 65 %, 2 %, 287 MiB sgc15: 38678, 587450 ms, 66 %, 2 %, 297 MiB sgc15: 38693, 587454 ms, 66 %, 2 %, 297 MiB sgc15: 38677, 587446 ms, 66 %, 2 %, 297 MiB sgc15: 38689, 587477 ms, 66 %, 2 %, 299 MiB sgc15: 1583, 588249 ms, 66 %, 2 %, 0 MiB sgc15: === GPU 3 === sgc15: pid, time [ms], gpu_utilization [%], mem_utilization [%], max_memory_usage [MiB] sgc15: 38710, 587341 ms, 65 %, 2 %, 297 MiB sgc15: 38707, 587344 ms, 65 %, 2 %, 299 MiB sgc15: 38695, 587344 ms, 65 %, 2 %, 297 MiB sgc15: 38709, 587348 ms, 65 %, 2 %, 299 MiB sgc15: 38699, 587361 ms, 65 %, 2 %, 297 MiB sgc15: 38667, 588210 ms, 65 %, 2 %, 287 MiB sgc15: 1583, 588245 ms, 66 %, 2 %, 0 MiB **************************************************************
The above report is generated by a 2-node GROMACS-2019 job which has 5 MPI ranks running on each GPU. (Note: the 0 MiB process is from NVML)
NVIDIA Datacenter GPU Manager
NVIDIA Management Library (NVML) only generates per-process GPU utilization report. A per-job report can be obtained by using NVIDIA Datacenter GPU Manager (DCGM). DCGM is installed on the SOSCPI GPU cluster but NOT enabled by default. User can launch DCGM as non-root with limited functionality.
- Note: DCGM job stats collecting may hurt performance especially for CPU intensive jobs. It is user's responsibility to understand the risk of downgraded performance.
To use DCGM to collect job stats, user needs to set SLURM Task Prolog/Epilog to insert command before/after the job execution.
Prolog Script
A SLRUM Task Prolog script (prolog.sh) should be used to launch DCGM daemon and start stats collection before job execution:
- User should have executable permission for prolog.sh (use command "chmod u+x prolog.sh" to add permission)
#!/bin/bash
if [ $SLURM_LOCALID = "0" ]; then #only 1 task per node runs the script
nv-hostengine & #launch DCGM daemon
dcgmi stats -e #enable stats recording
dcgmi stats -s $SLURM_JOB_ID #start collecting stats using SLURM job id as name
fi
Epilog Script
A SLURM Task Epilog script (epilog.sh) should be used to stop the stats recording and retrieve stats after job execution:
- User should have executable permission for epilog.sh (use command "chmod u+x epilog.sh" to add permission)
#!/bin/bash
if [ $SLURM_LOCALID = "0" ]; then #only 1 task per node runs the script
dcgmi stats -x $SLURM_JOB_ID #stop recording stats
dcgmi stats -j $SLURM_JOB_ID -v &> $SLURM_SUBMIT_DIR/dcgm-$SLURM_JOB_ID-`hostname`.out
#retrieve stats to file, e.g. dcgm-1234-sgc02.out
fi
Job Script Modification
SLURM job script should be modified to enable Task Prolog/Epilog scripts. SLURM_TASK_PROLOG and SLURM_TASK_EPILOG can be used to locate the scripts.
- srun is required to be used for launching job command. --mpi=pmi2 flag is needed for MPI jobs.
- For single-process job (e.g. Python), SLURM total task number must be 1.
A single-node Python example job:
#SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 export SLURM_TASK_PROLOG=$SLURM_SUBMIT_DIR/prolog.sh #path to prolog.sh file export SLURM_TASK_EPILOG=$SLURM_SUBMIT_DIR/epilog.sh #path to epilog.sh file srun --cpu_bind=none python code.py
- For multi-process job (e.g. MPI), SLURM total task number should be the same as the number of processes (e.g. number of MPI ranks)
An MPI GROMACS example job:
#SBATCH --nodes=1 #SBATCH --ntasks-per-node=20 export SLURM_TASK_PROLOG=$SLURM_SUBMIT_DIR/prolog.sh export SLURM_TASK_EPILOG=$SLURM_SUBMIT_DIR/epilog.sh srun --cpu_bind=none --mpi=pmi2 gmx_mpi mdrun -ntomp 2 -pin on -s nvt_1us.tpr
Example Output
- TensorFlow running on multiple GPUs:
Successfully retrieved statistics for job: 23681. +------------------------------------------------------------------------------+ | GPU ID: 0 | +====================================+=========================================+ |----- Execution Stats ------------+-----------------------------------------| | Start Time | Fri Nov 23 15:51:34 2018 | | End Time | Fri Nov 23 15:52:16 2018 | | Total Execution Time (sec) | 42.01 | | No. of Processes | 1 | +----- Performance Stats ----------+-----------------------------------------+ | Energy Consumed (Joules) | 3661 | | Power Usage (Watts) | Avg: 112.087, Max: 209.955, Min: 29.977 | | Max GPU Memory Used (bytes) | 16390291456 | | SM Clock (MHz) | Avg: 1278, Max: 1480, Min: 405 | | Memory Clock (MHz) | Avg: 715, Max: 715, Min: 715 | | SM Utilization (%) | Avg: 51, Max: 94, Min: 0 | | Memory Utilization (%) | Avg: 22, Max: 44, Min: 0 | | PCIe Rx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | | PCIe Tx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | +----- Event Stats ----------------+-----------------------------------------+ | Single Bit ECC Errors | 2 | | Double Bit ECC Errors | 0 | | PCIe Replay Warnings | 0 | | Critical XID Errors | 0 | +----- Slowdown Stats -------------+-----------------------------------------+ | Due to - Power (%) | 0 | | - Thermal (%) | 0 | | - Reliability (%) | Not Supported | | - Board Limit (%) | Not Supported | | - Low Utilization (%) | Not Supported | | - Sync Boost (%) | 0 | +-- Compute Process Utilization ---+-----------------------------------------+ | PID | 3865 | | Avg SM Utilization (%) | 34 | | Avg Memory Utilization (%) | 0 | +----- Overall Health -------------+-----------------------------------------+ | Overall Health | Healthy | +------------------------------------+-----------------------------------------+ +------------------------------------------------------------------------------+ | GPU ID: 1 | +====================================+=========================================+ |----- Execution Stats ------------+-----------------------------------------| | Start Time | Fri Nov 23 15:51:34 2018 | | End Time | Fri Nov 23 15:52:16 2018 | | Total Execution Time (sec) | 42.01 | | No. of Processes | 1 | +----- Performance Stats ----------+-----------------------------------------+ | Energy Consumed (Joules) | 3860 | | Power Usage (Watts) | Avg: 121.521, Max: 216.731, Min: 31.475 | | Max GPU Memory Used (bytes) | 16392388608 | | SM Clock (MHz) | Avg: 1278, Max: 1480, Min: 405 | | Memory Clock (MHz) | Avg: 715, Max: 715, Min: 715 | | SM Utilization (%) | Avg: 51, Max: 95, Min: 0 | | Memory Utilization (%) | Avg: 23, Max: 45, Min: 0 | | PCIe Rx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | | PCIe Tx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | +----- Event Stats ----------------+-----------------------------------------+ | Single Bit ECC Errors | 0 | | Double Bit ECC Errors | 0 | | PCIe Replay Warnings | 0 | | Critical XID Errors | 0 | +----- Slowdown Stats -------------+-----------------------------------------+ | Due to - Power (%) | 0 | | - Thermal (%) | 0 | | - Reliability (%) | Not Supported | | - Board Limit (%) | Not Supported | | - Low Utilization (%) | Not Supported | | - Sync Boost (%) | 0 | +-- Compute Process Utilization ---+-----------------------------------------+ | PID | 3866 | | Avg SM Utilization (%) | 33 | | Avg Memory Utilization (%) | 0 | +----- Overall Health -------------+-----------------------------------------+ | Overall Health | Healthy | +------------------------------------+-----------------------------------------+ +------------------------------------------------------------------------------+ | GPU ID: 2 | +====================================+=========================================+ |----- Execution Stats ------------+-----------------------------------------| | Start Time | Fri Nov 23 15:51:34 2018 | | End Time | Fri Nov 23 15:52:16 2018 | | Total Execution Time (sec) | 42.01 | | No. of Processes | 1 | +----- Performance Stats ----------+-----------------------------------------+ | Energy Consumed (Joules) | 3777 | | Power Usage (Watts) | Avg: 115.611, Max: 210.524, Min: 29.977 | | Max GPU Memory Used (bytes) | 16392388608 | | SM Clock (MHz) | Avg: 1278, Max: 1480, Min: 405 | | Memory Clock (MHz) | Avg: 715, Max: 715, Min: 715 | | SM Utilization (%) | Avg: 50, Max: 94, Min: 0 | | Memory Utilization (%) | Avg: 22, Max: 43, Min: 0 | | PCIe Rx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | | PCIe Tx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | +----- Event Stats ----------------+-----------------------------------------+ | Single Bit ECC Errors | 0 | | Double Bit ECC Errors | 0 | | PCIe Replay Warnings | 0 | | Critical XID Errors | 0 | +----- Slowdown Stats -------------+-----------------------------------------+ | Due to - Power (%) | 0 | | - Thermal (%) | 0 | | - Reliability (%) | Not Supported | | - Board Limit (%) | Not Supported | | - Low Utilization (%) | Not Supported | | - Sync Boost (%) | 0 | +-- Compute Process Utilization ---+-----------------------------------------+ | PID | 3867 | | Avg SM Utilization (%) | 33 | | Avg Memory Utilization (%) | 0 | +----- Overall Health -------------+-----------------------------------------+ | Overall Health | Healthy | +------------------------------------+-----------------------------------------+ +------------------------------------------------------------------------------+ | GPU ID: 3 | +====================================+=========================================+ |----- Execution Stats ------------+-----------------------------------------| | Start Time | Fri Nov 23 15:51:34 2018 | | End Time | Fri Nov 23 15:52:16 2018 | | Total Execution Time (sec) | 42.01 | | No. of Processes | 1 | +----- Performance Stats ----------+-----------------------------------------+ | Energy Consumed (Joules) | 3909 | | Power Usage (Watts) | Avg: 119.736, Max: 219.154, Min: 33.971 | | Max GPU Memory Used (bytes) | 16392388608 | | SM Clock (MHz) | Avg: 1278, Max: 1480, Min: 405 | | Memory Clock (MHz) | Avg: 715, Max: 715, Min: 715 | | SM Utilization (%) | Avg: 53, Max: 95, Min: 0 | | Memory Utilization (%) | Avg: 23, Max: 44, Min: 0 | | PCIe Rx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | | PCIe Tx Bandwidth (megabytes) | Avg: 0, Max: 0, Min: 0 | +----- Event Stats ----------------+-----------------------------------------+ | Single Bit ECC Errors | 0 | | Double Bit ECC Errors | 0 | | PCIe Replay Warnings | 0 | | Critical XID Errors | 0 | +----- Slowdown Stats -------------+-----------------------------------------+ | Due to - Power (%) | 0 | | - Thermal (%) | 0 | | - Reliability (%) | Not Supported | | - Board Limit (%) | Not Supported | | - Low Utilization (%) | Not Supported | | - Sync Boost (%) | 0 | +-- Compute Process Utilization ---+-----------------------------------------+ | PID | 3868 | | Avg SM Utilization (%) | 34 | | Avg Memory Utilization (%) | 0 | +----- Overall Health -------------+-----------------------------------------+ | Overall Health | Healthy | +------------------------------------+-----------------------------------------+
Documentation
- GPU Cluster Training slides: SOSCIP GPU Platform